1SUO
 
 | | Structure of mammalian cytochrome P450 2B4 with bound 4-(4-chlorophenyl)imidazole | | Descriptor: | 4-(4-CHLOROPHENYL)IMIDAZOLE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Scott, E.E, White, M.A, He, Y.A, Johnson, E.F, Stout, C.D, Halpert, J.R. | | Deposit date: | 2004-03-26 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of mammalian cytochrome P450 2B4 complexed with 4-(4-chlorophenyl)imidazole at 1.9 {angstrom} resolution: Insight into the range of P450 conformations and coordination of redox partner binding.
J.Biol.Chem., 279, 2004
|
|
2X5P
 
 | | Crystal structure of the Streptococcus pyogenes fibronectin binding protein Fbab-B | | Descriptor: | FIBRONECTIN BINDING PROTEIN | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-10 | | Release date: | 2010-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X4I
 
 | | ORF 114a from Sulfolobus islandicus rudivirus 1 | | Descriptor: | UNCHARACTERIZED PROTEIN 114 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, Naismith, J.H, White, M.F. | | Deposit date: | 2010-01-31 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X3G
 
 | | Crystal Structure of the hypothetical protein ORF119 from Sulfolobus islandicus rod-shaped virus 1 | | Descriptor: | SIRV1 HYPOTHETICAL PROTEIN ORF119 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-24 | | Release date: | 2010-07-21 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
2X3O
 
 | | Crystal Structure of the Hypothetical Protein PA0856 from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, GLYCEROL, HYPOTHETICAL PROTEIN PA0856 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X3M
 
 | | Crystal Structure of Hypothetical Protein ORF239 from Pyrobaculum Spherical Virus | | Descriptor: | HYPOTHETICAL PROTEIN ORF239 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-25 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X4L
 
 | | Crystal structure of DesE, a ferric-siderophore receptor protein from Streptomyces coelicolor | | Descriptor: | FERRIC-SIDEROPHORE RECEPTOR PROTEIN | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-01 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X7I
 
 | | Crystal structure of mevalonate kinase from methicillin-resistant Staphylococcus aureus MRSA252 | | Descriptor: | CHLORIDE ION, CITRIC ACID, MEVALONATE KINASE | | Authors: | Oke, M, Yan, X, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-27 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X5G
 
 | | Crystal structure of the ORF131L51M mutant from Sulfolobus islandicus rudivirus 1 | | Descriptor: | CHLORIDE ION, MALONATE ION, ORF 131 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, Naismith, J.H, White, M.F. | | Deposit date: | 2010-02-08 | | Release date: | 2010-07-21 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
2X3D
 
 | | Crystal Structure of SSo6206 from Sulfolobus solfataricus P2 | | Descriptor: | SSO6206 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, McMahon, S.A, McEwan, A.R, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-24 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Scottish Structural Proteomics Facility: targets, methods and outputs.
J. Struct. Funct. Genomics, 11, 2010
|
|
2X5H
 
 | | Crystal structure of the ORF131 L26M L51M double mutant from Sulfolobus islandicus rudivirus 1 | | Descriptor: | ORF 131, SULFATE ION | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, Naismith, J.H, White, M.F. | | Deposit date: | 2010-02-08 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
2W0M
 
 | | Crystal Structure of sso2452 from Sulfolobus solfataricus P2 | | Descriptor: | PYROPHOSPHATE 2-, SSO2452, ZINC ION | | Authors: | McRobbie, A, Carter, L, Johnson, K.A, Kerou, M, Liu, H, Mcmahon, S, Oke, M, Naismith, J.H, White, M.F. | | Deposit date: | 2008-08-19 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Characterisation of a Conserved Archaeal Rada Paralog with Antirecombinase Activity.
J.Mol.Biol., 389, 2009
|
|
2HWK
 
 | |
2XVO
 
 | | SSO1725, a protein involved in the CRISPR/Cas pathway | | Descriptor: | BETA-MERCAPTOETHANOL, SSO1725, SULFATE ION | | Authors: | Reeks, J, Liu, H, Naismith, J, White, M, McMahon, S. | | Deposit date: | 2010-10-26 | | Release date: | 2010-12-29 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure and Mechanism of the Cmr Complex for Crispr-Mediated Antiviral Immunity.
Mol.Cell, 45, 2012
|
|
2VL7
 
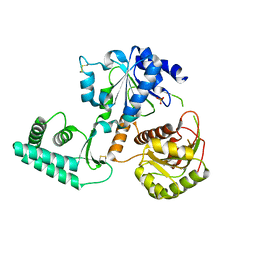 | | Structure of S. tokodaii Xpd4 | | Descriptor: | PHOSPHATE ION, XPD | | Authors: | Naismith, J.H, Johnson, K.A, Oke, M, McMahon, S.A, Liu, L, White, M.F, Zawadski, M, Carter, L.G. | | Deposit date: | 2008-01-08 | | Release date: | 2008-05-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the DNA Repair Helicase Xpd.
Cell(Cambridge,Mass.), 133, 2008
|
|
2VA8
 
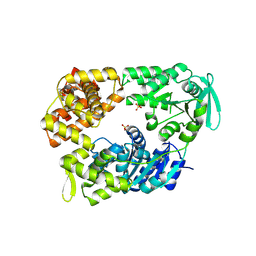 | | DNA Repair Helicase Hel308 | | Descriptor: | SKI2-TYPE HELICASE, SULFATE ION | | Authors: | Johnson, K.A, Richards, J, Liu, H, McMahon, S, Oke, M, Carter, L, Naismith, J.H, White, M.F. | | Deposit date: | 2007-08-30 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the DNA Repair Helicase Hel308 Reveals DNA Binding and Autoinhibitory Domains.
J.Biol.Chem., 283, 2008
|
|
6SCF
 
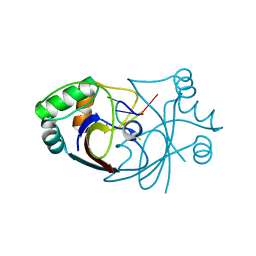 | | A viral anti-CRISPR subverts type III CRISPR immunity by rapid degradation of cyclic oligoadenylate | | Descriptor: | Uncharacterized protein, cyclic oligoadenylate | | Authors: | McMahon, S.A, Athukoralage, J.S, Graham, S, White, M.F, Gloster, T.M. | | Deposit date: | 2019-07-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An anti-CRISPR viral ring nuclease subverts type III CRISPR immunity.
Nature, 577, 2020
|
|
2IVY
 
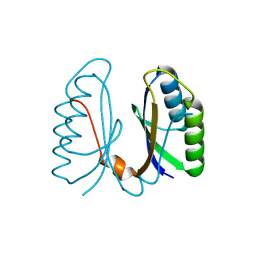 | | Crystal structure of hypothetical protein sso1404 from Sulfolobus solfataricus P2 | | Descriptor: | HYPOTHETICAL PROTEIN SSO1404 | | Authors: | Yan, X, Carter, L.G, Dorward, M, Liu, H, McMahon, S.A, Oke, M, Powers, H, White, M.F, Naismith, J.H. | | Deposit date: | 2006-06-22 | | Release date: | 2006-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
1Z66
 
 | | NMR solution structure of domain III of E-protein of tick-borne Langat flavivirus (no RDC restraints) | | Descriptor: | Major envelope protein E | | Authors: | Mukherjee, M, Dutta, K, White, M.A, Cowburn, D, Fox, R.O. | | Deposit date: | 2005-03-21 | | Release date: | 2006-03-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and backbone dynamics of domain III of the E protein of tick-borne Langat flavivirus suggests a potential site for molecular recognition.
Protein Sci., 15, 2006
|
|
1PXZ
 
 | | 1.7 Angstrom Crystal Structure of jun a 1, the major allergen from cedar pollen | | Descriptor: | Major pollen allergen Jun a 1 | | Authors: | Czerwinski, E.W, White, M.A, Midoro-Horiuti, T, Brooks, E.G, Goldblum, R.M. | | Deposit date: | 2003-07-07 | | Release date: | 2004-11-16 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Jun a 1, the major cedar pollen allergen from Juniperus ashei, reveals a parallel beta-helical core.
J.Biol.Chem., 280, 2005
|
|
2W02
 
 | | Co-complex Structure of Achromobactin Synthetase Protein D (AcsD) with ATP from Pectobacterium Chrysanthemi | | Descriptor: | ACSD, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Schmelz, S, McMahon, S.A, Kadi, N, Song, L, Oves-Costales, D, Oke, M, Liu, H, Johnson, K.A, Carter, L, White, M.F, Challis, G.L, Naismith, J.H. | | Deposit date: | 2008-08-08 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | AcsD catalyzes enantioselective citrate desymmetrization in siderophore biosynthesis.
Nat. Chem. Biol., 5, 2009
|
|
2X5D
 
 | | Crystal Structure of a probable aminotransferase from Pseudomonas aeruginosa | | Descriptor: | PROBABLE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-08 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
2X5T
 
 | | Crystal structure of ORF131 from Sulfolobus islandicus rudivirus 1 | | Descriptor: | MALONATE ION, ORF 131 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, Naismith, J.H, White, M.F. | | Deposit date: | 2010-02-10 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X7B
 
 | | Crystal structure of the N-terminal acetylase Ard1 from Sulfolobus solfataricus P2 | | Descriptor: | CHLORIDE ION, COENZYME A, N-ACETYLTRANSFERASE SSO0209 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, Mackay, D, White, M.F, Taylor, G.L, Naismith, J.H. | | Deposit date: | 2010-02-25 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
1H0Y
 
 | | Structure of Alba: an archaeal chromatin protein modulated by acetylation | | Descriptor: | DNA BINDING PROTEIN SSO10B, SULFATE ION | | Authors: | Wardleworth, B.N, Russell, R.J.M, Bell, S.D, Taylor, G.L, White, M.F. | | Deposit date: | 2002-07-01 | | Release date: | 2002-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Alba: An Archaeal Chromatin Protein Modulated by Acetylation
Embo J., 21, 2002
|
|
