1QL6
 
 | | THE CATALYTIC MECHANISM OF PHOSPHORYLASE KINASE PROBED BY MUTATIONAL STUDIES | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PHOSPHORYLASE KINASE, ... | | Authors: | Skamnaki, V.T, Owen, D.J, Noble, M.E.M, Lowe, E.D, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1999-08-24 | | Release date: | 1999-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catalytic Mechanism of Phosphorylase Kinase Probed by Mutational Studies.
Biochemistry, 38, 1999
|
|
2PHK
 
 | | THE CRYSTAL STRUCTURE OF A PHOSPHORYLASE KINASE PEPTIDE SUBSTRATE COMPLEX: KINASE SUBSTRATE RECOGNITION | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lowe, E.D, Noble, M.E.M, Skamnaki, V.T, Oikonomakos, N.G, Owen, D.J, Johnson, L.N. | | Deposit date: | 1998-06-18 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of a phosphorylase kinase peptide substrate complex: kinase substrate recognition.
EMBO J., 16, 1997
|
|
1NAF
 
 | |
2G30
 
 | | beta appendage of AP2 complexed with ARH peptide | | Descriptor: | 16-mer peptide from Low density lipoprotein receptor adapter protein 1, AP-2 complex subunit beta-1, peptide sequence AAF | | Authors: | Edeling, M.A, Collins, B.M, Traub, L.M, Owen, D.J. | | Deposit date: | 2006-02-17 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Switches Involving the AP-2 beta2 Appendage Regulate Endocytic Cargo Selection and Clathrin Coat Assembly
Dev.Cell, 10, 2006
|
|
4E47
 
 | | SET7/9 in complex with inhibitor (R)-(3-(3-cyanophenyl)-1-oxo-1-(pyrrolidin-1-yl)propan-2-yl)-1,2,3,4-tetrahydroisoquinoline-6- sulfonamide and S-adenosylmethionine | | Descriptor: | (R)-(3-(3-cyanophenyl)-1-oxo-1-(pyrrolidin-1-yl)propan-2-yl)-1,2,3,4-tetrahydroisoquinoline-6-sulfonamide, BETA-MERCAPTOETHANOL, Histone-lysine N-methyltransferase SETD7, ... | | Authors: | Walker, J.R, Ouyang, H, Dong, A, Fish, P, Cook, A, Barsyte, D, Vedadi, M, Tatlock, J, Owen, D, Bunnage, M, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-03-12 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Setd7 in Complex with Inhibitor and SAM
To be Published
|
|
4BX9
 
 | | Human Vps33A in complex with a fragment of human Vps16 | | Descriptor: | (2S)-2-hydroxybutanedioic acid, FORMIC ACID, MALONIC ACID, ... | | Authors: | Graham, S.C, Wartosch, L, Gray, S.R, Scourfield, E.J, Deane, J.E, Luzio, J.P, Owen, D.J. | | Deposit date: | 2013-07-09 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of Vps33A recruitment to the human HOPS complex by Vps16.
Proc. Natl. Acad. Sci. U.S.A., 110, 2013
|
|
4BX8
 
 | | Human Vps33A | | Descriptor: | CHLORIDE ION, VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 33A | | Authors: | Graham, S.C, Wartosch, L, Gray, S.R, Scourfield, E.J, Deane, J.E, Luzio, J.P, Owen, D.J. | | Deposit date: | 2013-07-09 | | Release date: | 2013-07-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Vps33A Recruitment to the Human Hops Complex by Vps16.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4E96
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor PFi-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-N-(3-methyl-2-oxo-1,2,3,4-tetrahydroquinazolin-6-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Fish, P, Bunnage, M, Owen, D, Knapp, S, Cook, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-03-20 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Identification of a chemical probe for bromo and extra C-terminal bromodomain inhibition through optimization of a fragment-derived hit.
J.Med.Chem., 55, 2012
|
|
1OM9
 
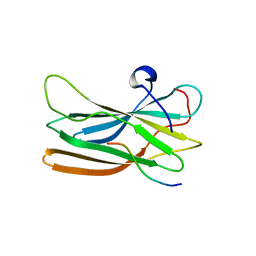 | | Structure of the GGA1-appendage in complex with the p56 binding peptide | | Descriptor: | 15-mer peptide fragment of p56, ADP-ribosylation factor binding protein GGA1 | | Authors: | Collins, B.M, Praefcke, G.J.K, Robinson, M.S, Owen, D.J. | | Deposit date: | 2003-02-25 | | Release date: | 2003-07-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for binding of accessory proteins by the appendage domain of GGAs
Nat.Struct.Biol., 10, 2003
|
|
4CCA
 
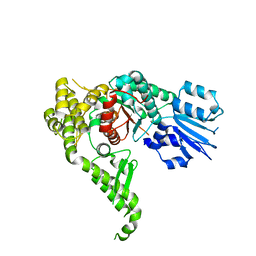 | | Structure of human Munc18-2 | | Descriptor: | CHLORIDE ION, SYNTAXIN-BINDING PROTEIN 2 | | Authors: | Hackmann, Y, Graham, S.C, Ehl, S, Hoening, S, Lehmberg, K, Arico, M, Owen, D.J, Griffiths, G.G. | | Deposit date: | 2013-10-21 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Syntaxin Binding Mechanism and Disease-Causing Mutations in Munc18-2
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HBY
 
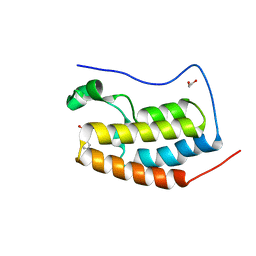 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazolin ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-2-oxo-N-phenyl-1,2,3,4-tetrahydroquinazoline-6-sulfonamide, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Fish, P.V, Bunnage, M.E, Cook, A.S, Owen, D.R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Identification of a Chemical Probe for Bromo and Extra C-Terminal Bromodomain Inhibition through Optimization of a Fragment-Derived Hit.
J.Med.Chem., 55, 2012
|
|
4HBV
 
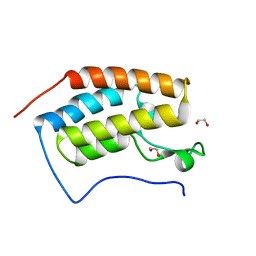 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazolin ligand | | Descriptor: | 1,2-ETHANEDIOL, 6-bromo-3-methyl-3,4-dihydroquinazolin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Fish, P.V, Bunnage, M.E, Cook, A.S, Owen, D.R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification of a Chemical Probe for Bromo and Extra C-Terminal Bromodomain Inhibition through Optimization of a Fragment-Derived Hit.
J.Med.Chem., 55, 2012
|
|
4HBW
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazoline ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-ethyl-3-methyl-2-oxo-1,2,3,4-tetrahydroquinazoline-6-sulfonamide, ... | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Fish, P.V, Bunnage, M.E, Cook, A.S, Owen, D.R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Identification of a Chemical Probe for Bromo and Extra C-Terminal Bromodomain Inhibition through Optimization of a Fragment-Derived Hit.
J.Med.Chem., 55, 2012
|
|
2RMK
 
 | | Rac1/PRK1 Complex | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Ras-related C3 botulinum toxin substrate 1, ... | | Authors: | Modha, R, Campbell, L.J, Nietlispach, D, Buhecha, H.R, Owen, D, Mott, H.R. | | Deposit date: | 2007-10-25 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Rac1 Polybasic Region Is Required for Interaction with Its Effector PRK1
J.Biol.Chem., 283, 2008
|
|
2BP5
 
 | | MU2 ADAPTIN SUBUNIT (AP50) OF AP2 ADAPTOR (SECOND DOMAIN), COMPLEXED WITH NON-CANONICAL INTERNALIZATION PEPTIDE VEDYEQGLSG | | Descriptor: | CLATHRIN COAT ASSEMBLY PROTEIN AP50, P2X PURINOCEPTOR 4 | | Authors: | Royle, S.J, Evans, P.R, Owen, D.J, Murrell-Lagnado, R.D. | | Deposit date: | 2005-04-18 | | Release date: | 2005-07-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Non-Canonical Yxxg-Phi Endocytic Motifs: Recognition by Ap2 and Preferential Utilization in P2X4 Receptors.
J.Cell.Sci, 118, 2005
|
|
1ZSG
 
 | | beta PIX-SH3 complexed with an atypical peptide from alpha-PAK | | Descriptor: | Rho guanine nucleotide exchange factor 7, Serine/threonine-protein kinase PAK 1 | | Authors: | Mott, H.R, Nietlispach, D, Evetts, K.A, Owen, D. | | Deposit date: | 2005-05-24 | | Release date: | 2005-08-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of the SH3 Domain of beta-PIX and Its Interaction with alpha-p21 Activated Kinase (PAK)
Biochemistry, 44, 2005
|
|
2QY7
 
 | | Crystal structure of human epsinR ENTH domain | | Descriptor: | Clathrin interactor 1 | | Authors: | Miller, S.E, Collins, B.M, McCoy, A.J, Robinson, M.S, Owen, D.J. | | Deposit date: | 2007-08-13 | | Release date: | 2007-11-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A SNARE-adaptor interaction is a new mode of cargo recognition in clathrin-coated vesicles.
Nature, 450, 2007
|
|
1Z2W
 
 | | Crystal structure of mouse Vps29 complexed with Mn2+ | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Vacuolar protein sorting 29 | | Authors: | Collins, B.M, Skinner, C.F, Watson, P.J, Seaman, M.N.J, Owen, D.J. | | Deposit date: | 2005-03-10 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Vps29 has a phosphoesterase fold that acts as a protein interaction scaffold for retromer assembly
NAT.STRUCT.MOL.BIOL., 12, 2005
|
|
4HBX
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazolin ligand | | Descriptor: | 3-methyl-6-(pyrrolidin-1-ylsulfonyl)-3,4-dihydroquinazolin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Fish, P.V, Bunnage, M.E, Cook, A.S, Owen, D.R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification of a Chemical Probe for Bromo and Extra C-Terminal Bromodomain Inhibition through Optimization of a Fragment-Derived Hit.
J.Med.Chem., 55, 2012
|
|
1Z2X
 
 | | Crystal structure of mouse Vps29 | | Descriptor: | Vacuolar protein sorting 29 | | Authors: | Collins, B.M, Skinner, C.F, Watson, P.J, Seaman, M.N.J, Owen, D.J. | | Deposit date: | 2005-03-10 | | Release date: | 2005-06-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Vps29 has a phosphoesterase fold that acts as a protein interaction scaffold for retromer assembly
NAT.STRUCT.MOL.BIOL., 12, 2005
|
|
1GYV
 
 | | Gamma-adaptin appendage domain from clathrin adaptor AP1, L762E mutant | | Descriptor: | ADAPTER-RELATED PROTEIN COMPLEX 1 GAMMA 1 SUBUNIT | | Authors: | Evans, P.R, Owen, D.J, McMahon, H.M, Kent, H.M. | | Deposit date: | 2002-04-30 | | Release date: | 2002-08-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Gamma-adaptin appendage domain: structure and binding site for Eps15 and gamma-synergin.
Structure, 10, 2002
|
|
1GYU
 
 | |
2QYW
 
 | | Crystal structure of mouse vti1b Habc domain | | Descriptor: | Vesicle transport through interaction with t-SNAREs 1B homolog | | Authors: | Miller, S.E, Collins, B.M, McCoy, A.J, Robinson, M.S, Owen, D.J. | | Deposit date: | 2007-08-15 | | Release date: | 2007-11-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A SNARE-adaptor interaction is a new mode of cargo recognition in clathrin-coated vesicles.
Nature, 450, 2007
|
|
1HK6
 
 | | Ral binding domain from Sec5 | | Descriptor: | EXOCYST COMPLEX COMPONENT SEC5 | | Authors: | Mott, H.R, Nietlispach, D, Hopkins, L.J, Mirey, G, Camonis, J.H, Owen, D. | | Deposit date: | 2003-03-05 | | Release date: | 2003-03-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the GTPase-binding domain of Sec5 and elucidation of its Ral binding site.
J. Biol. Chem., 278, 2003
|
|
2MT6
 
 | | Solution structure of the human ubiquitin conjugating enzyme Ube2w | | Descriptor: | Ubiquitin-conjugating enzyme E2 W | | Authors: | Vittal, V, Shi, L, Wenzel, D.M, Brzovic, P.S, Klevit, R.E. | | Deposit date: | 2014-08-14 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Intrinsic disorder drives N-terminal ubiquitination by Ube2w.
Nat.Chem.Biol., 11, 2015
|
|
