2RAB
 
 | | Structure of glutathione amide reductase from Chromatium gracile in complex with NAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, NICKEL (II) ION, ... | | Authors: | Van Petegem, F, De Vos, D, Savvides, S, Vergauwen, B, Van Beeumen, J. | | Deposit date: | 2007-09-14 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Understanding nicotinamide dinucleotide cofactor and substrate specificity in class I flavoprotein disulfide oxidoreductases: crystallographic analysis of a glutathione amide reductase.
J.Mol.Biol., 374, 2007
|
|
2R9Z
 
 | | Glutathione amide reductase from Chromatium gracile | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Glutathione amide reductase, ... | | Authors: | Van Petegem, F, Vergauwen, B, Savvides, S, De Vos, D, Van Beeumen, J. | | Deposit date: | 2007-09-14 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Understanding nicotinamide dinucleotide cofactor and substrate specificity in class I flavoprotein disulfide oxidoreductases: crystallographic analysis of a glutathione amide reductase.
J.Mol.Biol., 374, 2007
|
|
2C7E
 
 | | REVISED ATOMIC STRUCTURE FITTING INTO A GROEL(D398A)-ATP7 CRYO-EM MAP (EMD 1047) | | Descriptor: | 60 KDA CHAPERONIN, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ranson, N.A, Farr, G.W, Roseman, A.M, Gowen, B, Fenton, W.A, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2005-11-22 | | Release date: | 2006-02-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (14.9 Å) | | Cite: | ATP-Bound States of Groel Captured by Cryo-Electron Microscopy
Cell(Cambridge,Mass.), 107, 2001
|
|
1GR5
 
 | | Solution Structure of apo GroEL by Cryo-Electron microscopy | | Descriptor: | 60 KDA CHAPERONIN | | Authors: | Ranson, N.A, Farr, G.W, Roseman, A.M, Gowen, B, Fenton, W.A, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2001-12-14 | | Release date: | 2002-01-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | ATP-Bound States of Groel Captured by Cryo-Electron Microscopy.
Cell(Cambridge,Mass.), 107, 2001
|
|
3UCE
 
 | | Crystal structure of a small-chain dehydrogenase in complex with NADPH | | Descriptor: | Dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Buysschaert, G, Verstraete, K, Savvides, S, Vergauwen, B. | | Deposit date: | 2011-10-26 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural and biochemical characterization of an atypical short-chain dehydrogenase/reductase reveals an unusual cofactor preference.
Febs J., 280, 2013
|
|
3G9H
 
 | | Crystal structure of the C-terminal mu homology domain of Syp1 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Suppressor of yeast profilin deletion | | Authors: | Reider, A, Barker, S, Mishra, S, Im, Y.J, Maldonado-Baez, L, Hurley, J, Traub, L, Wendland, B. | | Deposit date: | 2009-02-13 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation.
Embo J., 28, 2009
|
|
3UCF
 
 | |
3G9G
 
 | | Crystal Structure of the N-terminal EFC/F-BAR domain of Syp1 | | Descriptor: | Suppressor of yeast profilin deletion | | Authors: | Reider, A, Barker, S, Mishra, S, Im, Y.J, Maldonado-Baez, L, Hurley, J, Traub, L, Wendland, B. | | Deposit date: | 2009-02-13 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation.
Embo J., 28, 2009
|
|
2O3J
 
 | | Structure of Caenorhabditis Elegans UDP-Glucose Dehydrogenase | | Descriptor: | GLYCEROL, UDP-glucose 6-dehydrogenase | | Authors: | Zhang, Y, Zhan, C, Patskovsky, Y, Ramagopal, U, Shi, W, Toro, R, Wengerter, B.C, Milst, S, Vidal, M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-12-01 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of Caenorhabditis Elegans Udp-Glucose Dehydrogenase
To be Published
|
|
1NO4
 
 | | Crystal Structure of the pre-assembly scaffolding protein gp7 from the double-stranded DNA bacteriophage phi29 | | Descriptor: | HEAD MORPHOGENESIS PROTEIN | | Authors: | Morais, M.C, Kanamaru, S, Badasso, M.O, Koti, J.S, Owen, B.A.L, McMurray, C.T, Anderson, D.L, Rossmann, M.G. | | Deposit date: | 2003-01-15 | | Release date: | 2003-07-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacteriophage f29 scaffolding protein gp7 before and after prohead assembly
Nat.Struct.Biol., 10, 2003
|
|
1DYL
 
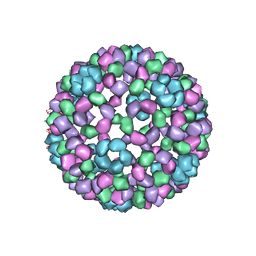 | | 9 ANGSTROM RESOLUTION CRYO-EM RECONSTRUCTION STRUCTURE OF SEMLIKI FOREST VIRUS (SFV) AND FITTING OF THE CAPSID PROTEIN STRUCTURE IN THE EM DENSITY | | Descriptor: | NUCLEOCAPSID PROTEIN | | Authors: | Mancini, E.J, Clarke, M, Gowen, B.E, Rutten, T, Fuller, S.D. | | Deposit date: | 2000-02-02 | | Release date: | 2000-08-18 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cryo-Electron Microscopy Reveals the Functional Organization of an Enveloped Virus, Semliki Forest Virus.
Mol.Cell, 5, 2000
|
|
1NOH
 
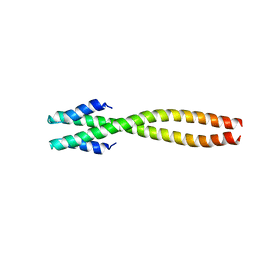 | | The structure of bacteriophage phi29 scaffolding protein gp7 after prohead assembly | | Descriptor: | HEAD MORPHOGENESIS PROTEIN | | Authors: | Morais, M.C, Kanamaru, S, Badasso, M.O, Koti, J.S, Owen, B.L, McMurray, C.T, L Anderson, D, Rossmann, M.G. | | Deposit date: | 2003-01-16 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bacteriophage f29 scaffolding protein gp7 before and after prohead assembly
Nat.Struct.Biol., 10, 2003
|
|
2H50
 
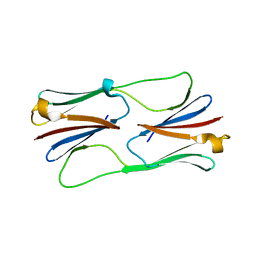 | | Multiple distinct assemblies reveal conformational flexibility in the small heat shock protein Hsp26 | | Descriptor: | small heat shock protein Hsp26 | | Authors: | White, H.E, Orlova, E.V, Chen, S, Wang, L, Ignatiou, A, Gowen, B, Stromer, T, Franzmann, T.M, Haslbeck, M, Buchner, J, Saibil, H.R. | | Deposit date: | 2006-05-25 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Multiple distinct assemblies reveal conformational flexibility in the small heat shock protein hsp26
Structure, 14, 2006
|
|
2H53
 
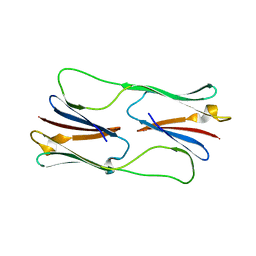 | | Multiple distinct assemblies reveal conformational flexibility in the small heat shock protein Hsp26 | | Descriptor: | small heat shock protein Hsp26 | | Authors: | White, H.E, Orlova, E.V, Chen, S, Wang, L, Ignatiou, A, Gowen, B, Stromer, T, Franzmann, T.M, Haslbeck, M, Buchner, J, Saibil, H.R. | | Deposit date: | 2006-05-25 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Multiple distinct assemblies reveal conformational flexibility in the small heat shock protein hsp26
Structure, 14, 2006
|
|
1GRU
 
 | | SOLUTION STRUCTURE OF GROES-ADP7-GROEL-ATP7 COMPLEX BY CRYO-EM | | Descriptor: | GROEL, GROES | | Authors: | Ranson, N.A, Farr, G.W, Roseman, A.M, Gowen, B, Fenton, W.A, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2001-12-16 | | Release date: | 2002-01-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | ATP-Bound States of Groel Captured by Cryo-Electron Microscopy.
Cell(Cambridge,Mass.), 107, 2001
|
|
1EJF
 
 | | CRYSTAL STRUCTURE OF THE HUMAN CO-CHAPERONE P23 | | Descriptor: | Prostaglandin E synthase 3, SULFATE ION | | Authors: | Weaver, A.J, Sullivan, W.P, Felts, S.J, Owen, B.A.L, Toft, D.O. | | Deposit date: | 2000-03-02 | | Release date: | 2000-06-19 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure and activity of human p23, a heat shock protein 90 co-chaperone.
J.Biol.Chem., 275, 2000
|
|
2HC9
 
 | | Structure of Caenorhabditis elegans leucine aminopeptidase-zinc complex (LAP1) | | Descriptor: | BICARBONATE ION, GLYCEROL, Leucine aminopeptidase 1, ... | | Authors: | Zhan, C, Patskovsky, Y, Wengerter, B.C, Ramagopal, U, Milstein, S, Vidal, M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-15 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure and Function of Caenorhabditis Elegans Leucine Aminopeptidase
To be Published
|
|
3LN7
 
 | |
3LN6
 
 | |
7K7G
 
 | | nucleosome and Gal4 complex | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit B, DNA (147-MER), Histone H2A.1, ... | | Authors: | Ruifang, G, Yawen, B. | | Deposit date: | 2020-09-22 | | Release date: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural and dynamic mechanisms of CBF3-guided centromeric nucleosome formation.
Nat Commun, 12, 2021
|
|
7K79
 
 | | CBF3 | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, Suppressor of kinetochore protein 1 | | Authors: | Ruifang, G, Yawen, B. | | Deposit date: | 2020-09-22 | | Release date: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural and dynamic mechanisms of CBF3-guided centromeric nucleosome formation.
Nat Commun, 12, 2021
|
|
7K78
 
 | | antibody and nucleosome complex | | Descriptor: | Cse4, DNA (136-MER), Histone H2A.1, ... | | Authors: | Ruifang, G, Yawen, B. | | Deposit date: | 2020-09-22 | | Release date: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and dynamic mechanisms of CBF3-guided centromeric nucleosome formation.
Nat Commun, 12, 2021
|
|
2MNG
 
 | | Apo Structure of human HCN4 CNBD solved by NMR | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Akimoto, M, Zhang, Z, Boulton, S, Selvaratnam, R, VanSchouwen, B, Gloyd, M, Accili, E.A, Lange, O.F, Melacini, G. | | Deposit date: | 2014-04-03 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A mechanism for the auto-inhibition of hyperpolarization-activated cyclic nucleotide-gated (HCN) channel opening and its relief by cAMP.
J.Biol.Chem., 289, 2014
|
|
