1DJM
 
 | | SOLUTION STRUCTURE OF BEF3-ACTIVATED CHEY FROM ESCHERICHIA COLI | | Descriptor: | CHEMOTAXIS PROTEIN Y | | Authors: | Cho, H.S, Lee, S.Y, Yan, D, Pan, X, Parkinson, J.S, Kustu, S, Wemmer, D.E, Pelton, J.G. | | Deposit date: | 1999-12-03 | | Release date: | 2000-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of activated CheY.
J.Mol.Biol., 297, 2000
|
|
1PUX
 
 | | NMR Solution Structure of BeF3-Activated Spo0F, 20 conformers | | Descriptor: | Sporulation initiation phosphotransferase F | | Authors: | Gardino, A.K, Volkman, B.F, Cho, H.S, Lee, S.Y, Wemmer, D.E, Kern, D. | | Deposit date: | 2003-06-25 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of BeF(3)(-)-activated Spo0F reveals the conformational switch in a phosphorelay system.
J.Mol.Biol., 331, 2003
|
|
2M20
 
 | | EGFR transmembrane - juxtamembrane (TM-JM) segment in bicelles: MD guided NMR refined structure. | | Descriptor: | Epidermal growth factor receptor | | Authors: | Endres, N.F, Das, R, Smith, A, Arkhipov, A, Kovacs, E, Huang, Y, Pelton, J.G, Shan, Y, Shaw, D.E, Wemmer, D.E, Groves, J.T, Kuriyan, J. | | Deposit date: | 2012-12-11 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Conformational Coupling across the Plasma Membrane in Activation of the EGF Receptor.
Cell(Cambridge,Mass.), 152, 2013
|
|
1I9F
 
 | | STRUCTURAL CHARACTERIZATION OF THE COMPLEX OF THE REV RESPONSE ELEMENT RNA WITH A SELECTED PEPTIDE | | Descriptor: | REV RESPONSE ELEMENT RNA, RSG-1.2 PEPTIDE | | Authors: | Zhang, Q, Harada, K, Cho, H.S, Frankel, A, Wemmer, D.E. | | Deposit date: | 2001-03-19 | | Release date: | 2001-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the complex of the Rev response element RNA with a selected peptide.
Chem.Biol., 8, 2001
|
|
1FQW
 
 | | CRYSTAL STRUCTURE OF ACTIVATED CHEY | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CHEMOTAXIS CHEY PROTEIN, MANGANESE (II) ION | | Authors: | Lee, S.Y, Cho, H.S, Pelton, J.G, Yan, D, Berry, E.A, Wemmer, D.E. | | Deposit date: | 2000-09-07 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of activated CheY. Comparison with other activated receiver domains.
J.Biol.Chem., 276, 2001
|
|
1PQQ
 
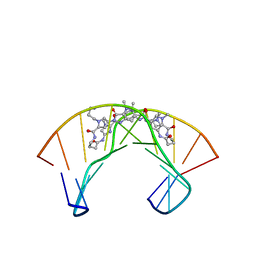 | | NMR Structure of a Cyclic Polyamide-DNA Complex | | Descriptor: | 45-(3-AMINOPROPYL)-5,11,22,28,34-PENTAMETHYL-3,9,15,20,26,32,38,43-OCTAOXO-2,5,8,14,19,22,25,28,31,34,37,42,45,48-TETRADECAAZA-11-AZONIAHEPTACYCLO[42.2.1.1~4,7~.1~10,13~.1~21,24~.1~27,30~.1~33,36~]DOPENTACONTA-1(46),4(52),6,10(51),12,21(50),23,27(49),29,33(48),35,44(47)-DODECAENE, 5'-D(*CP*GP*CP*TP*AP*AP*CP*AP*GP*GP*C)-3', 5'-D(*GP*CP*CP*TP*GP*TP*TP*AP*GP*CP*G)-3' | | Authors: | Zhang, Q, Dwyer, T.J, Tsui, V, Case, D.A, Cho, J, Dervan, P.B, Wemmer, D.E. | | Deposit date: | 2003-06-18 | | Release date: | 2004-06-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of a Cyclic Polyamide-DNA Complex.
J.Am.Chem.Soc., 126, 2004
|
|
1NY5
 
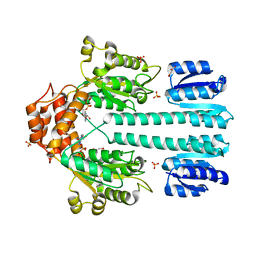 | | Crystal structure of sigm54 activator (AAA+ ATPase) in the inactive state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Lee, S.Y, de la Torre, A, Kustu, S, Nixon, B.T, Wemmer, D.E. | | Deposit date: | 2003-02-11 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulation of the transcriptional activator NtrC1: structural studies of the regulatory and AAA+ ATPase domains
Genes Dev., 17, 2003
|
|
2LAI
 
 | |
1IM1
 
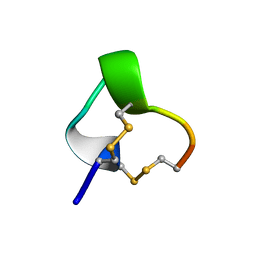 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1, 20 STRUCTURES | | Descriptor: | ALPHA-CONOTOXIN IM1 | | Authors: | Rogers, J.P, Luginbuhl, P, Shen, G.S, Mccabe, R.T, Stevens, R.C, Wemmer, D.E. | | Deposit date: | 1998-11-18 | | Release date: | 1999-06-15 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of alpha-conotoxin ImI and comparison to other conotoxins specific for neuronal nicotinic acetylcholine receptors.
Biochemistry, 38, 1999
|
|
1L7P
 
 | | SUBSTRATE BOUND PHOSPHOSERINE PHOSPHATASE COMPLEX STRUCTURE | | Descriptor: | PHOSPHATE ION, PHOSPHOSERINE, PHOSPHOSERINE PHOSPHATASE | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1L7N
 
 | | TRANSITION STATE ANALOGUE OF PHOSPHOSERINE PHOSPHATASE (ALUMINUM FLUORIDE COMPLEX) | | Descriptor: | ALUMINUM FLUORIDE, MAGNESIUM ION, PHOSPHOSERINE PHOSPHATASE, ... | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1L7O
 
 | | CRYSTAL STRUCTURE OF PHOSPHOSERINE PHOSPHATASE IN APO FORM | | Descriptor: | ACETIC ACID, PHOSPHOSERINE PHOSPHATASE, ZINC ION | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1DB6
 
 | | SOLUTION STRUCTURE OF THE DNA APTAMER 5'-CGACCAACGTGTCGCCTGGTCG-3' COMPLEXED WITH ARGININAMIDE | | Descriptor: | ARGININEAMIDE, DNA | | Authors: | Robertson, S.A, Harada, K, Frankel, A.D, Wemmer, D.E. | | Deposit date: | 1999-11-02 | | Release date: | 2000-02-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure determination and binding kinetics of a DNA aptamer-argininamide complex.
Biochemistry, 39, 2000
|
|
1NY6
 
 | | Crystal structure of sigm54 activator (AAA+ ATPase) in the active state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, transcriptional regulator (NtrC family) | | Authors: | Lee, S.Y, de la Torre, A, Kustu, S, Nixon, B.T, Wemmer, D.E. | | Deposit date: | 2003-02-11 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Regulation of the transcriptional activator NtrC1: structural studies of the regulatory and AAA+ ATPase domains
Genes Dev., 17, 2003
|
|
2O9L
 
 | | AMBER refined NMR Structure of the Sigma-54 RpoN Domain Bound to the-24 Promoter Element | | Descriptor: | 5'-D(*GP*AP*AP*AP*CP*GP*TP*GP*CP*CP*AP*AP*AP*A)-3', 5'-D(*TP*TP*TP*TP*GP*GP*CP*AP*CP*GP*TP*TP*TP*C)-3', RNA polymerase sigma factor RpoN | | Authors: | Doucleff, M, Pelton, J.G, Lee, P.S, Wemmer, D.E. | | Deposit date: | 2006-12-13 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA recognition by the alternative sigma-factor, sigma54.
J.Mol.Biol., 369, 2007
|
|
2O8K
 
 | | NMR Structure of the Sigma-54 RpoN Domain Bound to the-24 Promoter Element | | Descriptor: | 5'-D(*GP*AP*AP*AP*CP*GP*TP*GP*CP*CP*AP*AP*AP*A)-3', 5'-D(*TP*TP*TP*TP*GP*GP*CP*AP*CP*GP*TP*TP*TP*C)-3', RNA polymerase sigma factor RpoN | | Authors: | Doucleff, M, Pelton, J.G, Lee, P.S, Wemmer, D.E. | | Deposit date: | 2006-12-12 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA recognition by the alternative sigma-factor, sigma54.
J.Mol.Biol., 369, 2007
|
|
1L7M
 
 | | HIGH RESOLUTION LIGANDED STRUCTURE OF PHOSPHOSERINE PHOSPHATASE (PI COMPLEX) | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Phosphoserine Phosphatase | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-15 | | Release date: | 2002-04-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1J97
 
 | | Phospho-Aspartyl Intermediate Analogue of Phosphoserine phosphatase | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Phosphoserine Phosphatase | | Authors: | Cho, H, Wang, W, Kim, R, Yokota, H, Damo, S, Kim, S.-H, Wemmer, D, Kustu, S, Yan, D, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2001-05-24 | | Release date: | 2001-07-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | BeF(3)(-) acts as a phosphate analog in proteins phosphorylated on aspartate: structure of a BeF(3)(-) complex with phosphoserine phosphatase.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2PIS
 
 | | Efforts toward Expansion of the Genetic Alphabet: Structure and Replication of Unnatural Base Pairs | | Descriptor: | DNA (5'-D(*CP*GP*(CBR)P*GP*AP*AP*(FFD)P*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Matsuda, S, Fillo, J.D, Henry, A.A, Wilkins, S.J, Rai, P, Dwyer, T.J, Geierstanger, B.H, Wemmer, D.E, Schultz, P.G, Spraggon, G, Romesberg, F.E. | | Deposit date: | 2007-04-13 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Efforts toward expansion of the genetic alphabet: structure and replication of unnatural base pairs.
J.Am.Chem.Soc., 129, 2007
|
|
