7O1H
 
 | |
8R4E
 
 | |
8R6H
 
 | |
8RZX
 
 | |
8PSI
 
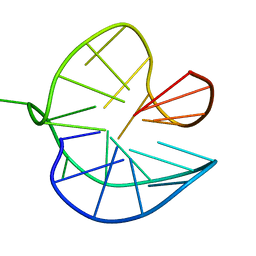 | |
8PSB
 
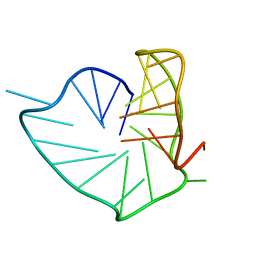 | | Three-layered parallel G-quadruplex with snapback loop from a G-rich sequence with five G-runs | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*TP*AP*GP*GP*GP*CP*GP*GP*CP*GP*GP*GP*GP*AP*CP*GP*GP*GP*T)-3') | | Authors: | Vianney, Y.M, Schroeder, N, Jana, J, Chojetzki, G, Weisz, K. | | Deposit date: | 2023-07-13 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-25 | | Method: | SOLUTION NMR | | Cite: | Showcasing Different G-Quadruplex Folds of a G-Rich Sequence: Between Rule-Based Prediction and Butterfly Effect.
J.Am.Chem.Soc., 145, 2023
|
|
6ZL2
 
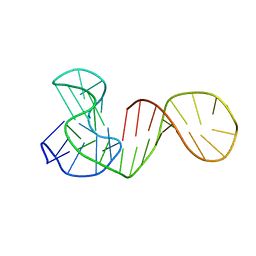 | |
6ZL9
 
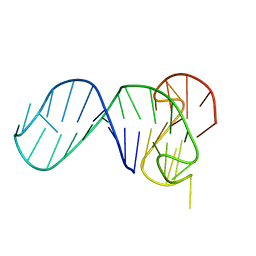 | |
6R9L
 
 | |
8RW2
 
 | |
6R9K
 
 | |
8PSE
 
 | |
2XQZ
 
 | | Neutron structure of the perdeuterated Toho-1 R274N R276N double mutant beta-lactamase | | Descriptor: | BETA-LACTAMSE TOHO-1 | | Authors: | Tomanicek, S.J, Wang, K.K, Weiss, K.L, Blakeley, M.P, Cooper, J, Chen, Y, Coates, L. | | Deposit date: | 2010-09-08 | | Release date: | 2010-12-22 | | Last modified: | 2024-05-08 | | Method: | NEUTRON DIFFRACTION (2.1 Å) | | Cite: | The Active Site Protonation States of Perdeuterated Toho-1 Beta-Lactamase Determined by Neutron Diffraction Support a Role for Glu166 as the General Base in Acylation.
FEBS Lett., 585, 2011
|
|
8R4W
 
 | |
8RAJ
 
 | | NMR structure of PKS docking domains | | Descriptor: | Beta-ketoacyl synthase, Trimethylamine monooxygenase | | Authors: | Scat, S, Weissman, K.J, Chagot, B. | | Deposit date: | 2023-12-01 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-17 | | Method: | SOLUTION NMR | | Cite: | Insights into docking in megasynthases from the investigation of the toblerol trans -AT polyketide synthase: many alpha-helical means to an end.
Rsc Chem Biol, 5, 2024
|
|
6C7A
 
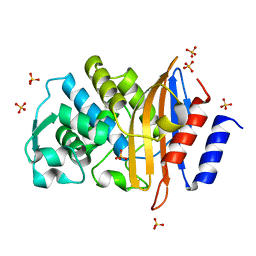 | | Conformational Changes in a Class A Beta lactamase that Prime it for Catalysis | | Descriptor: | Beta-lactamase Toho-1, SULFATE ION | | Authors: | Coates, L, Langan, P.S, Vandavasi, V.G, Cooper, S.J, Weiss, K.L, Ginell, S.L, Parks, J.M. | | Deposit date: | 2018-01-22 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Substrate Binding Induces Conformational Changes in a Class A Beta-lactamase That Prime It for Catalysis
Acs Catalysis, 8, 2018
|
|
8S1W
 
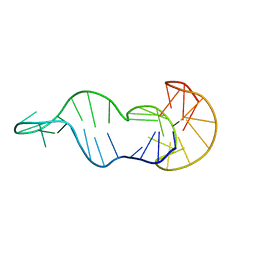 | |
2XR0
 
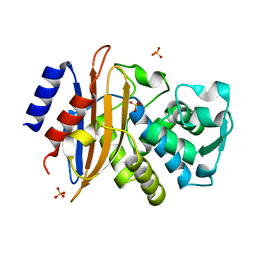 | | Room temperature X-ray structure of the perdeuterated Toho-1 R274N R276N double mutant beta-lactamase | | Descriptor: | SULFATE ION, TOHO-1 BETA-LACTAMASE | | Authors: | Tomanicek, S.J, Wang, K.K, Weiss, K.L, Blakeley, M.P, Cooper, J, Chen, Y, Coates, L. | | Deposit date: | 2010-09-08 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Active Site Protonation States of Perdeuterated Toho-1 Beta-Lactamase Determined by Neutron Diffraction Support a Role for Glu166 as the General Base in Acylation.
FEBS Lett., 585, 2011
|
|
4DCQ
 
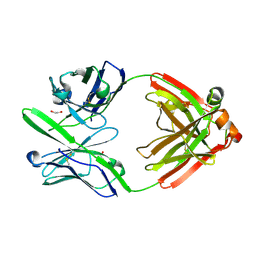 | | Crystal Structure of the Fab Fragment of 3B5H10, an Antibody-Specific for Extended Polyglutamine Repeats (orthorhombic form) | | Descriptor: | 1,2-ETHANEDIOL, 3B5H10 FAB Heavy Chain, 3B5H10 FAB Light Chain | | Authors: | Peters-Libeu, C.A, Tran, T, Finkbeiner, S, Weisgraber, K. | | Deposit date: | 2012-01-18 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Disease-associated polyglutamine stretches in monomeric huntingtin adopt a compact structure.
J.Mol.Biol., 421, 2012
|
|
6U58
 
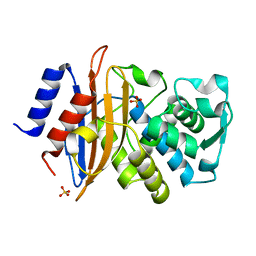 | | Toho1 Beta Lactamase Glu166Gln Mutant | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Langan, P.S, Sullivan, B, Weiss, K.L. | | Deposit date: | 2019-08-27 | | Release date: | 2020-02-19 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.9 Å), X-RAY DIFFRACTION | | Cite: | Probing the Role of the Conserved Residue Glu166 in a Class A Beta-Lactamase Using Neutron and X-ray Protein Crystallography
Acta Crystallogr.,Sect.D, 76, 2020
|
|
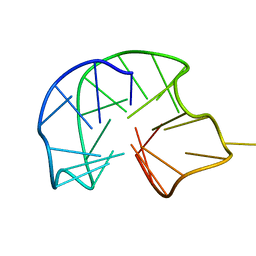
7ATZ
 
 | |
6ZTE
 
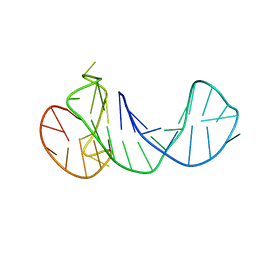 | |
6RS3
 
 | |
8A7Z
 
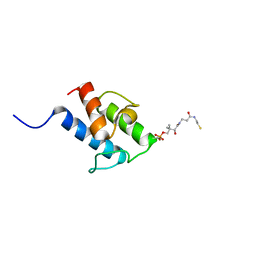 | | NMR structure of holo-acp | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Hybrid polyketide synthase-non ribosomal peptide synthetase | | Authors: | Collin, S, Weissman, K.J, Chagot, B, Gruez, A. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | SOLUTION NMR | | Cite: | Decrypting the programming of beta-methylation in virginiamycin M biosynthesis.
Nat Commun, 14, 2023
|
|
5KSC
 
 | | E166A/R274N/R276N Toho-1 Beta-lactamase aztreonam acyl-enzyme intermediate | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, Beta-lactamase Toho-1 | | Authors: | Vandavasi, V.G, Langan, P.S, Weiss, K, Parks, J.M, Cooper, J.B, Ginell, S.L, Coates, L. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2019-12-04 | | Method: | NEUTRON DIFFRACTION (2.1 Å) | | Cite: | Active-Site Protonation States in an Acyl-Enzyme Intermediate of a Class A beta-Lactamase with a Monobactam Substrate.
Antimicrob. Agents Chemother., 61, 2017
|
|
