6HN5
 
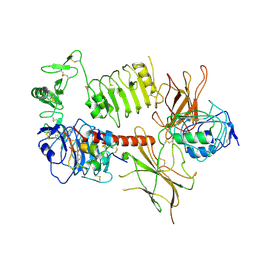 | | Leucine-zippered human insulin receptor ectodomain with single bound insulin - "upper" membrane-distal part | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin, ... | | Authors: | Weis, F, Menting, J.G, Margetts, M.B, Chan, S.J, Xu, Y, Tennagels, N, Wohlfart, P, Langer, T, Mueller, C.W, Dreyer, M.K, Lawrence, M.C. | | Deposit date: | 2018-09-14 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The signalling conformation of the insulin receptor ectodomain.
Nat Commun, 9, 2018
|
|
6SAE
 
 | | Cryo-EM structure of TMV in water | | Descriptor: | Capsid protein, MAGNESIUM ION, RNA (5'-R(P*GP*AP*A)-3') | | Authors: | Weis, F, Beckers, M, Sachse, C. | | Deposit date: | 2019-07-16 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Elucidation of the viral disassembly switch of tobacco mosaic virus.
Embo Rep., 20, 2019
|
|
6SAG
 
 | | Cryo-EM structure of TMV with Ca2+ at low pH | | Descriptor: | CALCIUM ION, Capsid protein, MAGNESIUM ION, ... | | Authors: | Weis, F, Beckers, M, Sachse, C. | | Deposit date: | 2019-07-16 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Elucidation of the viral disassembly switch of tobacco mosaic virus.
Embo Rep., 20, 2019
|
|
3IYQ
 
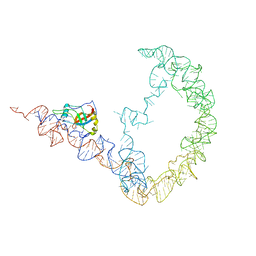 | | tmRNA-SmpB: a journey to the center of the bacterial ribosome | | Descriptor: | SsrA-binding protein, tmRNA | | Authors: | Weis, F, Bron, P, Giudice, E, Rolland, J.P, Thomas, D, Felden, B, Gillet, R. | | Deposit date: | 2010-04-16 | | Release date: | 2010-10-20 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | tmRNA-SmpB: a journey to the centre of the bacterial ribosome.
Embo J., 29, 2010
|
|
3IYR
 
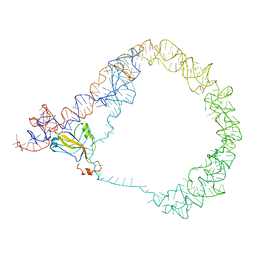 | | tmRNA-SmpB: a journey to the center of the bacterial ribosome | | Descriptor: | SsrA-binding protein, tmRNA | | Authors: | Weis, F, Bron, P, Giudice, E, Rolland, J.P, Thomas, D, Felden, B, Gillet, R. | | Deposit date: | 2010-04-16 | | Release date: | 2010-10-20 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | tmRNA-SmpB: a journey to the centre of the bacterial ribosome.
Embo J., 29, 2010
|
|
6HN4
 
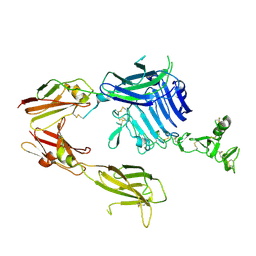 | | Leucine-zippered human insulin receptor ectodomain with single bound insulin - "lower" membrane-proximal part | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin receptor,Insulin receptor,General control protein GCN4 | | Authors: | Weis, F, Menting, J.G, Margetts, M.B, Chan, S.J, Xu, Y, Tennagels, N, Wohlfart, P, Langer, T, Mueller, C.W, Dreyer, M.K, Lawrence, M.C. | | Deposit date: | 2018-09-14 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The signalling conformation of the insulin receptor ectodomain.
Nat Commun, 9, 2018
|
|
5ANC
 
 | | Mechanism of eIF6 release from the nascent 60S ribosomal subunit | | Descriptor: | 26S RIBOSOMAL RNA, 60S ACIDIC RIBOSOMAL PROTEIN P0, 60S RIBOSOMAL PROTEIN L10, ... | | Authors: | Weis, F, Giudice, E, Churcher, M, Jin, L, Hilcenko, C, Wong, C.C, Traynor, D, Kay, R.R, Warren, A.J. | | Deposit date: | 2015-09-06 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mechanism of Eif6 Release from the Nascent 60S Ribosomal Subunit
Nat.Struct.Mol.Biol., 22, 2015
|
|
5ANB
 
 | | Mechanism of eIF6 release from the nascent 60S ribosomal subunit | | Descriptor: | 26S RIBOSOMAL RNA, 60S ACIDIC RIBOSOMAL PROTEIN P0, 60S RIBOSOMAL PROTEIN L10, ... | | Authors: | Weis, F, Giudice, E, Churcher, M, Jin, L, Hilcenko, C, Wong, C.C, Traynor, D, Kay, R.R, Warren, A.J. | | Deposit date: | 2015-09-06 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Mechanism of Eif6 Release from the Nascent 60S Ribosomal Subunit
Nat.Struct.Mol.Biol., 22, 2015
|
|
5AN9
 
 | | Mechanism of eIF6 release from the nascent 60S ribosomal subunit | | Descriptor: | 26S RIBOSOMAL RNA, 60S ACIDIC RIBOSOMAL PROTEIN P0, 60S RIBOSOMAL PROTEIN L10, ... | | Authors: | Weis, F, Giudice, E, Churcher, M, Jin, L, Hilcenko, C, Wong, C.C, Traynor, D, Kay, R.R, Warren, A.J. | | Deposit date: | 2015-09-06 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of Eif6 Release from the Nascent 60S Ribosomal Subunit
Nat.Struct.Mol.Biol., 22, 2015
|
|
8CDP
 
 | | Cryo-EM structure of the RESC1-RESC2 complex | | Descriptor: | Guide_RNA_associated_protein_-_putative, Mitochondrial guide RNA binding complex subunit 2 | | Authors: | Dolce, L.G, Weis, F, Kowalinski, E. | | Deposit date: | 2023-01-31 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for guide RNA selection by the RESC1-RESC2 complex.
Nucleic Acids Res., 51, 2023
|
|
6EZO
 
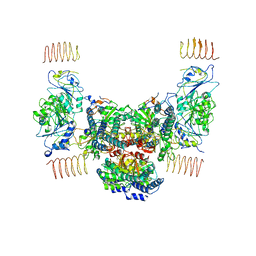 | | Eukaryotic initiation factor EIF2B in complex with ISRIB | | Descriptor: | 2-(4-chloranylphenoxy)-~{N}-[4-[2-(4-chloranylphenoxy)ethanoylamino]cyclohexyl]ethanamide, Human eukaryotic initiation factor EIF2B epsilon subunits, Translation initiation factor eIF-2B subunit alpha, ... | | Authors: | Faille, A, Weis, F, Zyryanova, A, Warren, A.J, Ron, D. | | Deposit date: | 2017-11-16 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Binding of ISRIB reveals a regulatory site in the nucleotide exchange factor eIF2B.
Science, 359, 2018
|
|
8AW3
 
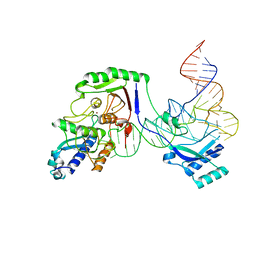 | | Cryo-EM structure of the Tb ADAT2/3 deaminase in complex with tRNA | | Descriptor: | Deaminase, putative, RNA (75-MER), ... | | Authors: | Dolce, L.G, Tengo, L, Weis, F, Kowalinski, E. | | Deposit date: | 2022-08-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for sequence-independent substrate selection by eukaryotic wobble base tRNA deaminase ADAT2/3.
Nat Commun, 13, 2022
|
|
6QPW
 
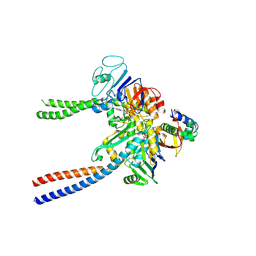 | | Structural basis of cohesin ring opening | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Sister chromatid cohesion protein 1, ... | | Authors: | Panne, D, Muir, K.W, Li, Y, Weis, F. | | Deposit date: | 2019-02-15 | | Release date: | 2020-02-05 | | Last modified: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structure of the cohesin ATPase elucidates the mechanism of SMC-kleisin ring opening.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6HKO
 
 | | Yeast RNA polymerase I elongation complex bound to nucleotide analog GMPCPP | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Tafur, L, Sadian, Y, Weis, F, Muller, C.W. | | Deposit date: | 2018-09-07 | | Release date: | 2019-04-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | The cryo-EM structure of a 12-subunit variant of RNA polymerase I reveals dissociation of the A49-A34.5 heterodimer and rearrangement of subunit A12.2.
Elife, 8, 2019
|
|
6HLR
 
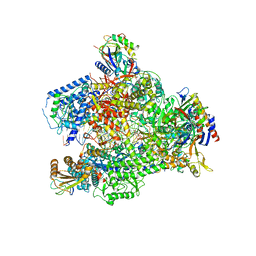 | | Yeast RNA polymerase I elongation complex bound to nucleotide analog GMPCPP (core focused) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Tafur, L, Sadian, Y, Weis, F, Muller, C.W. | | Deposit date: | 2018-09-11 | | Release date: | 2019-04-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The cryo-EM structure of a 12-subunit variant of RNA polymerase I reveals dissociation of the A49-A34.5 heterodimer and rearrangement of subunit A12.2.
Elife, 8, 2019
|
|
6HLS
 
 | | Yeast apo RNA polymerase I* | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Tafur, L, Sadian, Y, Weis, F, Muller, C.W. | | Deposit date: | 2018-09-11 | | Release date: | 2019-04-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | The cryo-EM structure of a 12-subunit variant of RNA polymerase I reveals dissociation of the A49-A34.5 heterodimer and rearrangement of subunit A12.2.
Elife, 8, 2019
|
|
6HLQ
 
 | | Yeast RNA polymerase I* elongation complex bound to nucleotide analog GMPCPP | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Tafur, L, Sadian, Y, Weis, F, Muller, C.W. | | Deposit date: | 2018-09-11 | | Release date: | 2019-04-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The cryo-EM structure of a 12-subunit variant of RNA polymerase I reveals dissociation of the A49-A34.5 heterodimer and rearrangement of subunit A12.2.
Elife, 8, 2019
|
|
8QFS
 
 | | Cryo-EM structure of SidH from Legionella pneumophila | | Descriptor: | Elongation factor Tu, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sharma, R, Weis, F, Bhogaraju, S. | | Deposit date: | 2023-09-04 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for the toxicity of Legionella pneumophila effector SidH.
Nat Commun, 14, 2023
|
|
8S0B
 
 | | H. sapiens MCM bound to double stranded DNA and ORC6 as part of the MCM-ORC complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (45-mer), DNA replication licensing factor MCM2, ... | | Authors: | Greiwe, J.F, Weissmann, F, Diffley, J.F.X, Costa, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | MCM Double Hexamer Loading Visualised with Human Proteins
Nature, 2024
|
|
8S0F
 
 | | H. sapiens OC1M bound to double stranded DNA | | Descriptor: | DNA (39-mer), DNA replication factor Cdt1, DNA replication licensing factor MCM2, ... | | Authors: | Greiwe, J.F, Weissmann, F, Diffley, J.F.X, Costa, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | MCM Double Hexamer Loading Visualised with Human Proteins
Nature, 2024
|
|
8S0D
 
 | | H. sapiens MCM bound to double stranded DNA and ORC1-6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (58-mer), DNA replication licensing factor MCM2, ... | | Authors: | Greiwe, J.F, Weissmann, F, Diffley, J.F.X, Costa, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | MCM Double Hexamer Loading Visualised with Human Proteins
Nature, 2024
|
|
8S0E
 
 | | H. sapiens OCCM bound to double stranded DNA | | Descriptor: | Cell division control protein 6 homolog, DNA (39-mer), DNA replication factor Cdt1, ... | | Authors: | Greiwe, J.F, Weissmann, F, Diffley, J.F.X, Costa, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | MCM Double Hexamer Loading Visualised with Human Proteins
Nature, 2024
|
|
8S0A
 
 | | H. sapiens MCM2-7 hexamer bound to double stranded DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (22-mer), ... | | Authors: | Greiwe, J.F, Weissmann, F, Diffley, J.F.X, Costa, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | MCM Double Hexamer Loading Visualised with Human Proteins
Nature, 2024
|
|
8S09
 
 | | H. sapiens MCM2-7 double hexamer bound to double stranded DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (45-mer), ... | | Authors: | Greiwe, J.F, Weissmann, F, Diffley, J.F.X, Costa, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | MCM Double Hexamer Loading Visualised with Human Proteins
Nature, 2024
|
|
8S0C
 
 | | H. sapiens ORC1-5 bound to double stranded DNA as part of the MCM-ORC complex | | Descriptor: | DNA (26-mer), Isoform 2 of Origin recognition complex subunit 3, MAGNESIUM ION, ... | | Authors: | Greiwe, J.F, Weissmann, F, Diffley, J.F.X, Costa, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | MCM Double Hexamer Loading Visualised with Human Proteins
Nature, 2024
|
|
