7VS3
 
 | | The crystal structure of rat calcium-dependent activator protein for secretion (CAPS) C2PH | | Descriptor: | Calcium-dependent secretion activator 1, SULFATE ION | | Authors: | Zhou, H, Wei, Z.Q, Zhang, L, Ren, Y.J, Ma, C. | | Deposit date: | 2021-10-25 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | The C 2 and PH domains of CAPS constitute an effective PI(4,5)P2-binding unit essential for Ca 2+ -regulated exocytosis.
Structure, 31, 2023
|
|
6D26
 
 | | Crystal structure of the prostaglandin D2 receptor CRTH2 with fevipiprant | | Descriptor: | OLEIC ACID, Prostaglandin D2 receptor 2, Endolysin chimera, ... | | Authors: | Wang, L, Yao, D, Deepak, K, Liu, H, Gong, W, Fan, H, Wei, Z, Zhang, C. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Structures of the Human PGD2Receptor CRTH2 Reveal Novel Mechanisms for Ligand Recognition.
Mol. Cell, 72, 2018
|
|
6D27
 
 | | Crystal structure of the prostaglandin D2 receptor CRTH2 with CAY10471 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, OLEIC ACID, ... | | Authors: | Wang, L, Yao, D, Deepak, K, Liu, H, Gong, W, Fan, H, Wei, Z, Zhang, C. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.738 Å) | | Cite: | Structures of the Human PGD2Receptor CRTH2 Reveal Novel Mechanisms for Ligand Recognition.
Mol. Cell, 72, 2018
|
|
5Y4E
 
 | | Crystal Structure of AnkB Ankyrin Repeats R8-14 in complex with autoinhibition segment AI-b | | Descriptor: | Ankyrin-2,Ankyrin-2, GLYCEROL, SULFATE ION | | Authors: | Chen, K, Li, J, Wang, C, Wei, Z, Zhang, M. | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Autoinhibition of ankyrin-B/G membrane target bindings by intrinsically disordered segments from the tail regions.
Elife, 6, 2017
|
|
5Y4D
 
 | | Crystal Structure of AnkB Ankyrin Repeats in Complex with AnkR/AnkB Chimeric Autoinhibition Segment | | Descriptor: | Ankyrin-1,Ankyrin-2,Ankyrin-2, SULFATE ION | | Authors: | Chen, K, Li, J, Wang, C, Wei, Z, Zhang, M. | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Autoinhibition of ankyrin-B/G membrane target bindings by intrinsically disordered segments from the tail regions.
Elife, 6, 2017
|
|
5Y4F
 
 | | Crystal Structure of AnkB Ankyrin Repeats R13-24 in complex with autoinhibition segment AI-c | | Descriptor: | ACETATE ION, Ankyrin-2, CALCIUM ION | | Authors: | Chen, K, Li, J, Wang, C, Wei, Z, Zhang, M. | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Autoinhibition of ankyrin-B/G membrane target bindings by intrinsically disordered segments from the tail regions.
Elife, 6, 2017
|
|
7FH7
 
 | | Friedel-Crafts alkylation enzyme CylK mutant Y37F | | Descriptor: | 5-[(2S,7R)-7-fluoranyl-2-methyl-undecyl]benzene-1,3-diol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, H.Q, Wei, Z, Xiang, Z. | | Deposit date: | 2021-07-29 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural Basis for the Friedel-Crafts Alkylation in Cylindrocyclophane Biosynthesis
ACS Catal., 12, 2022
|
|
7FH8
 
 | | Friedel-Crafts alkylation enzyme CylK mutant H391A | | Descriptor: | 2-[(5S,10S)-11-[3,5-bis(oxidanyl)phenyl]-10-methyl-undecan-5-yl]-5-[(2S,7R)-7-fluoranyl-2-methyl-undecyl]benzene-1,3-diol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, H.Q, Wei, Z, Xiang, Z. | | Deposit date: | 2021-07-29 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural Basis for the Friedel-Crafts Alkylation in Cylindrocyclophane Biosynthesis
ACS Catal., 12, 2022
|
|
7D2T
 
 | | Crystal structure of Rsu1/PINCH1_LIM45C complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, LIM and senescent cell antigen-like-containing domain protein 1, ... | | Authors: | Yang, H, Wei, Z, Yu, C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Complex structures of Rsu1 and PINCH1 reveal a regulatory mechanism of the ILK/PINCH/Parvin complex for F-actin dynamics.
Elife, 10, 2021
|
|
7D2S
 
 | | Crystal structure of Rsu1/PINCH1_LIM5C complex | | Descriptor: | GLYCEROL, LIM and senescent cell antigen-like-containing domain protein 1, Ras suppressor protein 1, ... | | Authors: | Yang, H, Wei, Z, Cong, Y. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Complex structures of Rsu1 and PINCH1 reveal a regulatory mechanism of the ILK/PINCH/Parvin complex for F-actin dynamics.
Elife, 10, 2021
|
|
7D2U
 
 | | Crystal structure of Rsu1/PINCH1_LIM45C complex | | Descriptor: | GLYCEROL, LIM and senescent cell antigen-like-containing domain protein 1, MALONATE ION, ... | | Authors: | Yang, H, Wei, Z, Cong, Y. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Complex structures of Rsu1 and PINCH1 reveal a regulatory mechanism of the ILK/PINCH/Parvin complex for F-actin dynamics.
Elife, 10, 2021
|
|
7D2E
 
 | | Tetrameric coiled-coil structure of liprin-alpha2_H3 | | Descriptor: | Liprin-alpha-2 | | Authors: | Liang, M, Wei, Z. | | Deposit date: | 2020-09-16 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Oligomerized liprin-alpha promotes phase separation of ELKS for compartmentalization of presynaptic active zone proteins.
Cell Rep, 34, 2021
|
|
7D2G
 
 | | Coiled-coil structure of liprin-alpha2_H2delC | | Descriptor: | GLYCEROL, Liprin-alpha-2 | | Authors: | Liang, M, Wei, Z. | | Deposit date: | 2020-09-16 | | Release date: | 2021-04-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Oligomerized liprin-alpha promotes phase separation of ELKS for compartmentalization of presynaptic active zone proteins.
Cell Rep, 34, 2021
|
|
7D2H
 
 | | Tetrameric coiled-coil structure of liprin-alpha2_H2 | | Descriptor: | GLYCEROL, Liprin-alpha-2 | | Authors: | Liang, M, Wei, Z. | | Deposit date: | 2020-09-16 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oligomerized liprin-alpha promotes phase separation of ELKS for compartmentalization of presynaptic active zone proteins.
Cell Rep, 34, 2021
|
|
7DDX
 
 | | Crystal structure of KANK1 S1179F mutant in complex wtih eIF4A1 | | Descriptor: | Eukaryotic initiation factor 4A-I, GLYCEROL, KN motif and ankyrin repeat domains 1, ... | | Authors: | Pan, W, Xu, Y, Wei, Z. | | Deposit date: | 2020-10-30 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nephrotic-syndrome-associated mutation of KANK2 induces pathologic binding competition with physiological interactor KIF21A.
J.Biol.Chem., 297, 2021
|
|
6IUH
 
 | | Crystal structure of GIT1 PBD domain in complex with Liprin-alpha2 | | Descriptor: | ARF GTPase-activating protein GIT1, IODIDE ION, Liprin-alpha-2 | | Authors: | Liang, M, Wei, Z. | | Deposit date: | 2018-11-28 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the target-binding mode of the G protein-coupled receptor kinase-interacting protein in the regulation of focal adhesion dynamics.
J. Biol. Chem., 294, 2019
|
|
6IXQ
 
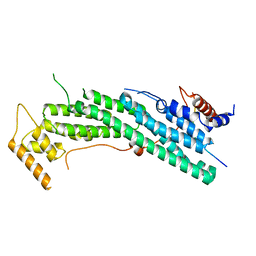 | | Structure of Myo2-GTD in complex with Smy1 | | Descriptor: | Kinesin-related protein SMY1, Myosin-2 | | Authors: | Tang, K, Wei, Z. | | Deposit date: | 2018-12-11 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural mechanism for versatile cargo recognition by the yeast class V myosin Myo2.
J.Biol.Chem., 294, 2019
|
|
6IXR
 
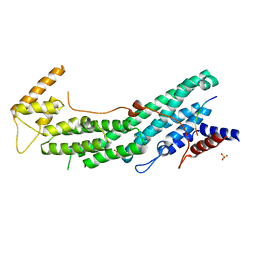 | | Structure of Myo2-GTD in complex with Inp2 | | Descriptor: | Inheritance of peroxisomes protein 2, Myosin-2, SULFATE ION | | Authors: | Tang, K, Wei, Z. | | Deposit date: | 2018-12-11 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.854 Å) | | Cite: | Structural mechanism for versatile cargo recognition by the yeast class V myosin Myo2.
J.Biol.Chem., 294, 2019
|
|
6IXO
 
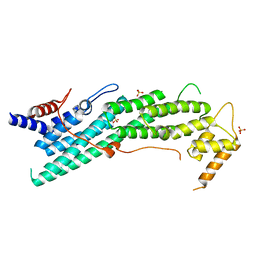 | | Apo structure of Myo2-GTD | | Descriptor: | CHLORIDE ION, Myosin-2, SULFATE ION | | Authors: | Tang, K, Wei, Z. | | Deposit date: | 2018-12-11 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural mechanism for versatile cargo recognition by the yeast class V myosin Myo2.
J.Biol.Chem., 294, 2019
|
|
6IXP
 
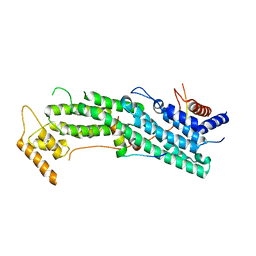 | | Structure of Myo2-GTD in complex with Mmr1 | | Descriptor: | Mitochondrial MYO2 receptor-related protein 1, Myosin-2 | | Authors: | Tang, K, Wei, Z. | | Deposit date: | 2018-12-11 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.733 Å) | | Cite: | Structural mechanism for versatile cargo recognition by the yeast class V myosin Myo2.
J.Biol.Chem., 294, 2019
|
|
6KR4
 
 | | Crystal structure of the liprin-alpha3_SAM123/LAR_D1D2 complex | | Descriptor: | HEXAETHYLENE GLYCOL, Liprin-alpha-3, PHOSPHATE ION, ... | | Authors: | Xie, X, Liang, M, Luo, L, Wei, Z. | | Deposit date: | 2019-08-20 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of liprin-alpha-promoted LAR-RPTP clustering for modulation of phosphatase activity.
Nat Commun, 11, 2020
|
|
4AFX
 
 | | Crystal structure of the reactive loop cleaved ZPI in I2 space group | | Descriptor: | CALCIUM ION, PHOSPHATE ION, PROTEIN Z DEPENDENT PROTEASE INHIBITOR, ... | | Authors: | Zhou, A, Yan, Y, Wei, Z. | | Deposit date: | 2012-01-23 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Basis for Catalytic Activation of Protein Z-Dependent Protease Inhibitor (Zpi) by Protein Z.
Blood, 120, 2012
|
|
4AJU
 
 | | Crystal structure of the reactive loop cleaved ZPI in P41 space group | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN Z-DEPENDENT PROTEASE INHIBITOR, SULFATE ION | | Authors: | Zhou, A, Yan, Y, Wei, Z. | | Deposit date: | 2012-02-19 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Basis for Catalytic Activation of Protein Z-Dependent Protease Inhibitor (Zpi) by Protein Z.
Blood, 120, 2012
|
|
6F4U
 
 | |
2HNK
 
 | | Crystal structure of SAM-dependent O-methyltransferase from pathogenic bacterium Leptospira interrogans | | Descriptor: | DI(HYDROXYETHYL)ETHER, S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent O-methyltransferase, ... | | Authors: | Hou, X, Wei, Z, Gong, W. | | Deposit date: | 2006-07-13 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of SAM-dependent O-methyltransferase from pathogenic bacterium Leptospira interrogans.
J.Struct.Biol., 159, 2007
|
|
