5A93
 
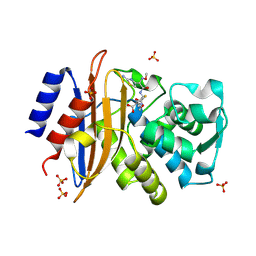 | | 293K Joint X-ray Neutron with Cefotaxime: EXPLORING THE MECHANISM OF BETA-LACTAM RING PROTONATION IN THE CLASS A BETA-LACTAMASE ACYLATION MECHANISM USING NEUTRON AND X-RAY CRYSTALLOGRAPHY | | Descriptor: | BETA-LACTAMASE CTX-M-97, CEFOTAXIME, C3' cleaved, ... | | Authors: | Vandavasi, V.G, Weiss, K.L, Cooper, J.B, Erskine, P.T, Tomanicek, S.J, Ostermann, A, Schrader, T.E, Ginell, S.L, Coates, L. | | Deposit date: | 2015-07-17 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-09 | | Method: | NEUTRON DIFFRACTION (1.598 Å), X-RAY DIFFRACTION | | Cite: | Exploring the Mechanism of Beta-Lactam Ring Protonation in the Class a Beta-Lactamase Acylation Mechanism Using Neutron and X-Ray Crystallography.
J.Med.Chem., 59, 2016
|
|
5A90
 
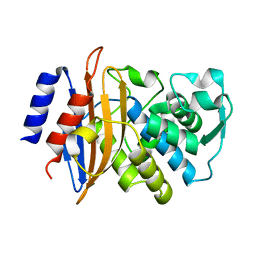 | | 100K Neutron Ligand Free: Exploring the Mechanism of beta-Lactam Ring Protonation in the Class A beta-lactamase Acylation Mechanism Using Neutron and X-ray Crystallography | | Descriptor: | BETA-LACTAMASE CTX-M-97 | | Authors: | Vandavasi, V.G, Weiss, K.L, Cooper, J.B, Erskine, P.T, Tomanicek, S.J, Ostermann, A, Schrader, T.E, Ginell, S.L, Coates, L. | | Deposit date: | 2015-07-17 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-08 | | Method: | NEUTRON DIFFRACTION (1.7 Å) | | Cite: | Exploring the Mechanism of Beta-Lactam Ring Protonation in the Class a Beta-Lactamase Acylation Mechanism Using Neutron and X-Ray Crystallography.
J.Med.Chem., 59, 2016
|
|
5A91
 
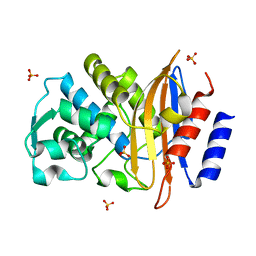 | | 15K X-ray ligand free: Exploring the Mechanism of beta-Lactam Ring Protonation in the Class A beta-lactamase Acylation Mechanism Using Neutron and X-ray Crystallography | | Descriptor: | Beta-lactamase Toho-1, SULFATE ION | | Authors: | Vandavasi, V.G, Weiss, K.L, Cooper, J.B, Erskine, P.T, Tomanicek, S.J, Ostermann, A, Schrader, T.E, Ginell, S.L, Coates, L. | | Deposit date: | 2015-07-17 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Exploring the Mechanism of Beta-Lactam Ring Protonation in the Class a Beta-Lactamase Acylation Mechanism Using Neutron and X-Ray Crystallography.
J.Med.Chem., 59, 2016
|
|
5A92
 
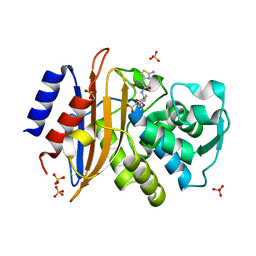 | | 15K X-ray structure with Cefotaxime: Exploring the Mechanism of beta- Lactam Ring Protonation in the Class A beta-lactamase Acylation Mechanism Using Neutron and X-ray Crystallography | | Descriptor: | BETA-LACTAMASE CTX-M-97, CEFOTAXIME, C3' cleaved, ... | | Authors: | Vandavasi, V.G, Weiss, K.L, Cooper, J.B, Erskine, P.T, Tomanicek, S.J, Ostermann, A, Schrader, T.E, Ginell, S.L, Coates, L. | | Deposit date: | 2015-07-17 | | Release date: | 2015-12-16 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Exploring the Mechanism of Beta-Lactam Ring Protonation in the Class a Beta-Lactamase Acylation Mechanism Using Neutron and X-Ray Crystallography.
J.Med.Chem., 59, 2016
|
|
1E4R
 
 | | Solution structure of the mouse defensin mBD-8 | | Descriptor: | Beta-defensin 8 | | Authors: | Bauer, F, Schweimer, K, Kluver, E, Adermann, K, Forssmann, W.G, Roesch, P, Sticht, H. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2018-06-20 | | Method: | SOLUTION NMR | | Cite: | Structure determination of human and murine beta-defensins reveals structural conservation in the absence of significant sequence similarity.
Protein Sci., 10, 2001
|
|
1H4B
 
 | | SOLUTION STRUCTURE OF THE BIRCH POLLEN ALLERGEN BET V 4 | | Descriptor: | CALCIUM ION, POLCALCIN BET V 4 | | Authors: | Neudecker, P, Nerkamp, J, Eisenmann, A, Lauber, T, Lehmann, K, Schweimer, K, Roesch, P. | | Deposit date: | 2003-02-26 | | Release date: | 2004-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Dynamics, and Hydrodynamics of the Calcium-Bound Cross-Reactive Birch Pollen Allergen Bet V 4 Reveal a Canonical Monomeric Two EF-Hand Assembly with a Regulatory Function
J.Mol.Biol., 336, 2004
|
|
4BD0
 
 | | X-ray structure of a perdeuterated Toho-1 R274N R276N double mutant Beta-lactamase in complex with a fully deuterated boronic acid (BZB) | | Descriptor: | BENZO[B]THIOPHENE-2-BORONIC ACID, BETA-LACTAMASE TOHO-1, SULFATE ION | | Authors: | Tomanicek, S.J, Weiss, K.L, Standaert, R.F, Ostermann, A, Schrader, T.E, Ng, J.D, Coates, L. | | Deposit date: | 2012-10-04 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.207 Å) | | Cite: | Neutron and X-Ray Crystal Structures of a Perdeuterated Enzyme Inhibitor Complex Reveal the Catalytic Proton Network of the Toho-1 Beta-Lactamase for the Acylation Reaction.
J.Biol.Chem., 288, 2013
|
|
1WCN
 
 | |
1WCL
 
 | |
1IOJ
 
 | | HUMAN APOLIPOPROTEIN C-I, NMR, 18 STRUCTURES | | Descriptor: | APOC-I | | Authors: | Rozek, A, Sparrow, J.T, Weisgraber, K.H, Cushley, R.J. | | Deposit date: | 1998-05-12 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Conformation of human apolipoprotein C-I in a lipid-mimetic environment determined by CD and NMR spectroscopy.
Biochemistry, 38, 1999
|
|
1W1F
 
 | | SH3 DOMAIN OF HUMAN LYN TYROSINE KINASE | | Descriptor: | TYROSINE-PROTEIN KINASE LYN | | Authors: | Bauer, F, Schweimer, K, Hoffmann, S, Roesch, P, Sticht, H. | | Deposit date: | 2004-06-17 | | Release date: | 2005-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Lyn-SH3 Domain in Complex with a Herpesviral Protein Reveals an Extended Recognition Motif that Enhances Binding Affinity.
Protein Sci., 14, 2005
|
|
4BD1
 
 | | Neutron structure of a perdeuterated Toho-1 R274N R276N double mutant Beta-lactamase in complex with a fully deuterated boronic acid (BZB) | | Descriptor: | BENZO[B]THIOPHENE-2-BORONIC ACID, TOHO-1 BETA-LACTAMASE | | Authors: | Tomanicek, S.J, Weiss, K.L, Standaert, R.F, Ostermann, A, Schrader, T.E, Ng, J.D, Coates, L. | | Deposit date: | 2012-10-04 | | Release date: | 2013-01-16 | | Last modified: | 2017-03-22 | | Method: | NEUTRON DIFFRACTION (2.002 Å) | | Cite: | Neutron and X-Ray Crystal Structures of a Perdeuterated Enzyme Inhibitor Complex Reveal the Catalytic Proton Network of the Toho-1 Beta-Lactamase for the Acylation Reaction.
J.Biol.Chem., 288, 2013
|
|
1WA7
 
 | | SH3 DOMAIN OF HUMAN LYN TYROSINE KINASE IN COMPLEX WITH A HERPESVIRAL LIGAND | | Descriptor: | HYPOTHETICAL 28.7 KDA PROTEIN IN DHFR 3'REGION (ORF1), TYROSINE-PROTEIN KINASE LYN | | Authors: | Bauer, F, Schweimer, K, Hoffmann, S, Roesch, P, Sticht, H. | | Deposit date: | 2004-10-25 | | Release date: | 2005-07-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Investigation of the Binding of a Herpesviral Protein to the SH3 Domain of Tyrosine Kinase Lck.
Biochemistry, 41, 2002
|
|
1KOA
 
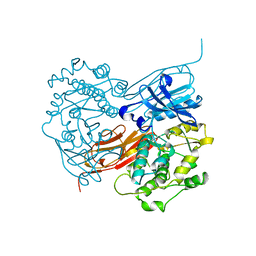 | | TWITCHIN KINASE FRAGMENT (C.ELEGANS), AUTOREGULATED PROTEIN KINASE AND IMMUNOGLOBULIN DOMAINS | | Descriptor: | TWITCHIN | | Authors: | Kobe, B, Heierhorst, J, Feil, S.C, Parker, M.W, Benian, G.M, Weiss, K.R, Kemp, B.E. | | Deposit date: | 1996-06-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Giant protein kinases: domain interactions and structural basis of autoregulation.
EMBO J., 15, 1996
|
|
1KOB
 
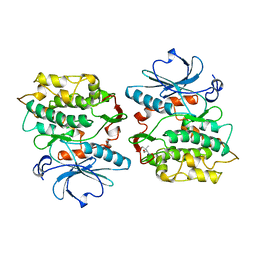 | | TWITCHIN KINASE FRAGMENT (APLYSIA), AUTOREGULATED PROTEIN KINASE DOMAIN | | Descriptor: | TWITCHIN, VALINE | | Authors: | Kobe, B, Heierhorst, J, Feil, S.C, Parker, M.W, Benian, G.M, Weiss, K.R, Kemp, B.E. | | Deposit date: | 1996-06-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Giant protein kinases: domain interactions and structural basis of autoregulation.
EMBO J., 15, 1996
|
|
1PZQ
 
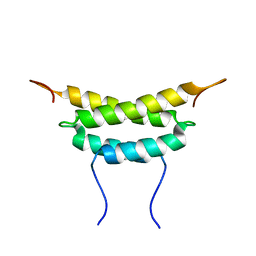 | | Structure of fused docking domains from the erythromycin polyketide synthase (DEBS), a model for the interaction between DEBS 2 and DEBS 3: The A domain | | Descriptor: | Erythronolide synthase | | Authors: | Broadhurst, R.W, Nietlispach, D, Wheatcroft, M.P, Leadlay, P.F, Weissman, K.J. | | Deposit date: | 2003-07-14 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of docking domains in modular polyketide synthases.
Chem.Biol., 10, 2003
|
|
1YA9
 
 | | Crystal Structure of the 22kDa N-Terminal Fragment of Mouse Apolipoprotein E | | Descriptor: | Apolipoprotein E | | Authors: | Peters-Libeu, C.A, Rutenber, E, Newhouse, Y, Hatters, D.M, Weisgraber, K.H. | | Deposit date: | 2004-12-17 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Engineering conformational destabilization into mouse apolipoprotein E. A model for a unique property of human apolipoprotein E4
J.Biol.Chem., 280, 2005
|
|
4GQB
 
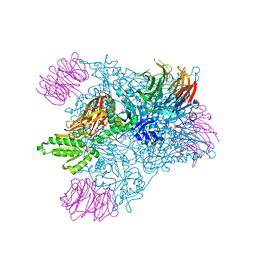 | | Crystal Structure of the human PRMT5:MEP50 Complex | | Descriptor: | (2S,5S,6E)-2,5-diamino-6-[(3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxydihydrofuran-2(3H)-ylidene]hexanoic acid, Histone H4 peptide, Methylosome protein 50, ... | | Authors: | Antonysamy, S, Bonday, Z, Campbell, R, Doyle, B, Druzina, Z, Gheyi, T, Han, B, Jungheim, L.N, Qian, Y, Rauch, C, Russell, M, Sauder, J.M, Wasserman, S.R, Weichert, K, Willard, F.S, Zhang, A, Emtage, S. | | Deposit date: | 2012-08-22 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of the human PRMT5:MEP50 complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1PZR
 
 | | Structure of fused docking domains from the erythromycin polyketide synthase (DEBS), a model for the interaction between DEBS2 and DEBS3: the B domain | | Descriptor: | Erythronolide synthase | | Authors: | Broadhurst, R.W, Nietlispach, D, Wheatcroft, M.P, Leadlay, P.F, Weissman, K.J. | | Deposit date: | 2003-07-14 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of docking domains in modular polyketide synthases.
Chem.Biol., 10, 2003
|
|
1E10
 
 | | [2Fe-2S]-Ferredoxin from Halobacterium salinarum | | Descriptor: | ACETYL GROUP, FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Marg, B.-L, Schweimer, K, Oesterhelt, D, Roesch, P, Sticht, H. | | Deposit date: | 2000-04-12 | | Release date: | 2001-04-09 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | A Two-Alpha-Helix Extra Domain Mediates the Halophilic Character of a Plant-Type Ferredoxin from Halophilic Archaea.
Biochemistry, 44, 2005
|
|
3SS2
 
 | | Neutron structure of perdeuterated rubredoxin using 48 hours 3rd pass data | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Munshi, P, Chung, C.-L, Blakeley, M.P, Weiss, K.L, Myles, D.A.A, Meilleur, F. | | Deposit date: | 2011-07-07 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (1.75 Å) | | Cite: | Rapid visualization of hydrogen positions in protein neutron crystallographic structures.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2VCD
 
 | | Solution structure of the FKBP-domain of Legionella pneumophila Mip in complex with rapamycin | | Descriptor: | Outer membrane protein MIP, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Ceymann, A, Horstmann, M, Ehses, P, Schweimer, K, Paschke, A.-K, Fischer, G, Roesch, P, Faber, C. | | Deposit date: | 2007-09-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Legionella pneumophila Mip-rapamycin complex.
BMC Struct. Biol., 8, 2008
|
|
2HCC
 
 | | SOLUTION STRUCTURE OF THE HUMAN CHEMOKINE HCC-2, NMR, 30 STRUCTURES | | Descriptor: | HUMAN CHEMOKINE HCC-2 | | Authors: | Sticht, H, Escher, S.E, Schweimer, K, Forssmann, W.G, Roesch, P, Adermann, K. | | Deposit date: | 1998-07-03 | | Release date: | 1999-07-13 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human CC chemokine 2: A monomeric representative of the CC chemokine subtype.
Biochemistry, 38, 1999
|
|
2K7H
 
 | | NMR solution structure of soybean allergen Gly m 4 | | Descriptor: | Stress-induced protein SAM22 | | Authors: | Berkner, H, Neudecker, P, Mittag, D, Ballmer-Weber, B.K, Schweimer, K, Vieths, S, Roesch, P. | | Deposit date: | 2008-08-11 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Cross-reactivity of pollen and food allergens: soybean Gly m 4 is a member of the Bet v 1 superfamily and closely resembles yellow lupine proteins
Biosci.Rep., 29, 2009
|
|
2KTT
 
 | | Solution Structure of a Covalently Bound Pyrrolo[2,1-c][1,4]benzodiazepine-Benzimidazole Hybrid to a 10mer DNA Duplex | | Descriptor: | (11aS)-7-methoxy-8-(3-{4-[6-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]phenoxy}propoxy)-1,2,3,10,11,11a-hexahydro-5H-pyrrolo[2,1-c][1,4]benzodiazepin-5-one, 5'-D(*AP*AP*CP*AP*AP*TP*TP*GP*TP*T)-3' | | Authors: | Rettig, M, Weingarth, M, Langel, W, Kamal, A, Kumar, P.P, Weisz, K. | | Deposit date: | 2010-02-08 | | Release date: | 2010-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a covalently bound pyrrolo[2,1-c][1,4]benzodiazepine-benzimidazole hybrid to a 10mer DNA duplex.
Biochemistry, 48, 2009
|
|
