2JNZ
 
 | |
2KWP
 
 | |
8R6K
 
 | | Crystal structure of Candida glabrata Bdf1 bromodomain 1 bound to a phenyltriazine ligand | | Descriptor: | 6-methyl-~{N}-[(5-methylfuran-2-yl)methyl]-3-(4-methylphenyl)-1,2,4-triazin-5-amine, Candida glabrata strain CBS138 chromosome C complete sequence, PHOSPHATE ION, ... | | Authors: | Petosa, C, Wei, K, McKenna, C.E, Govin, J. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Humanized Candida and NanoBiT Assays Expedite Discovery of Bdf1 Bromodomain Inhibitors with Antifungal Potential against invasive Candida infection
Advanced Science, 2024
|
|
6B85
 
 | | Crystal structure of transmembrane protein TMHC4_R | | Descriptor: | TMHC4_R | | Authors: | Lu, P, DiMaio, F, Min, D, Bowie, J, Wei, K.Y, Baker, D. | | Deposit date: | 2017-10-05 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.889 Å) | | Cite: | Accurate computational design of multipass transmembrane proteins.
Science, 359, 2018
|
|
6B87
 
 | | Crystal structure of transmembrane protein TMHC2_E | | Descriptor: | TMHC2_E | | Authors: | Lu, P, DiMaio, F, Min, D, Wei, K.Y, Bowie, J, Baker, D. | | Deposit date: | 2017-10-05 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.947 Å) | | Cite: | Accurate computational design of multipass transmembrane proteins.
Science, 359, 2018
|
|
8R6M
 
 | | Crystal structure of Candida glabrata Bdf1 bromodomain 2 bound to I-BET151 | | Descriptor: | 7-(3,5-DIMETHYL-1,2-OXAZOL-4-YL)-8-METHOXY-1-[(1R)-1-(PYRIDIN-2-YL)ETHYL]-1H,2H,3H-IMIDAZO[4,5-C]QUINOLIN-2-ONE, Candida glabrata strain CBS138 chromosome C complete sequence, GLYCEROL, ... | | Authors: | Petosa, C, Wei, K, McKenna, C.E, Govin, J. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Humanized Candida and NanoBiT Assays Expedite Discovery of Bdf1 Bromodomain Inhibitors with Antifungal Potential against invasive Candida infection
Advanced Science, 2024
|
|
8R6L
 
 | | Crystal structure of Candida glabrata Bdf1 bromodomain 2 in the unbound state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Candida glabrata strain CBS138 chromosome C complete sequence, ... | | Authors: | Petosa, C, Wei, K, McKenna, C.E, Govin, J. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Humanized Candida and NanoBiT Assays Expedite Discovery of Bdf1 Bromodomain Inhibitors with Antifungal Potential against invasive Candida infection
Advanced Science, 2024
|
|
8R6I
 
 | |
8R6J
 
 | | Crystal structure of Candida glabrata Bdf1 bromodomain 1 bound to a pyrazole ligand | | Descriptor: | 2-methyl-~{N}-[[5-(3-thiophen-2-yl-1,2,4-oxadiazol-5-yl)thiophen-2-yl]methyl]pyrazole-3-carboxamide, Candida glabrata strain CBS138 chromosome C complete sequence, SULFATE ION | | Authors: | Petosa, C, Wei, K, McKenna, C.E, Govin, J. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Humanized Candida and NanoBiT Assays Expedite Discovery of Bdf1 Bromodomain Inhibitors with Antifungal Potential against invasive Candida infection
Advanced Science, 2024
|
|
8R6N
 
 | | Crystal structure of Candida glabrata Bdf1 bromodomain 2 bound to a pyridoindole ligand | | Descriptor: | 2-ethanoyl-~{N}-(4-morpholin-4-ylphenyl)-1,3,4,5-tetrahydropyrido[4,3-b]indole-8-carboxamide, Candida glabrata strain CBS138 chromosome C complete sequence | | Authors: | Petosa, C, Wei, K, McKenna, C.E, Govin, J. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Humanized Candida and NanoBiT Assays Expedite Discovery of Bdf1 Bromodomain Inhibitors with Antifungal Potential against invasive Candida infection
Advanced Science, 2024
|
|
4GD6
 
 | | SHV-1 beta-lactamase in complex with penam sulfone SA1-204 | | Descriptor: | (3R)-N-(2-formylindolizin-3-yl)-4-[(phenylacetyl)oxy]-3-sulfino-D-valine, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | van den Akker, F, Wei, K. | | Deposit date: | 2012-07-31 | | Release date: | 2013-07-31 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structures of SHV-1 beta-lactamase with penem and penam sulfone inhibitors that form cyclic intermediates stabilized by carbonyl conjugation
Plos One, 7, 2012
|
|
1XIP
 
 | |
7PER
 
 | | Model of the inner ring of the human nuclear pore complex | | Descriptor: | Nuclear pore complex protein Nup155, Nuclear pore complex protein Nup205, Nuclear pore complex protein Nup93, ... | | Authors: | Schuller, A.P, Wojtynek, M, Mankus, D, Tatli, M, Kronenberg-Tenga, R, Regmi, S.G, Dasso, M, Weis, K, Medalia, O, Schwartz, T.U. | | Deposit date: | 2021-08-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | The cellular environment shapes the nuclear pore complex architecture.
Nature, 598, 2021
|
|
7PEQ
 
 | | Model of the outer rings of the human nuclear pore complex | | Descriptor: | Nuclear pore complex protein Nup107, Nuclear pore complex protein Nup133, Nuclear pore complex protein Nup160, ... | | Authors: | Schuller, A.P, Wojtynek, M, Mankus, D, Tatli, M, Kronenberg-Tenga, R, Regmi, S.G, Dasso, M, Weis, K, Medalia, O, Schwartz, T.U. | | Deposit date: | 2021-08-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | The cellular environment shapes the nuclear pore complex architecture.
Nature, 598, 2021
|
|
3GFP
 
 | | Structure of the C-terminal domain of the DEAD-box protein Dbp5 | | Descriptor: | DEAD box protein 5 | | Authors: | Erzberger, J.P, Dossani, Z.Y, Weirich, C.S, Weis, K, Berger, J.M. | | Deposit date: | 2009-02-27 | | Release date: | 2009-09-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the C-terminus of the mRNA export factor Dbp5 reveals the interaction surface for the ATPase activator Gle1
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1QGR
 
 | | STRUCTURE OF IMPORTIN BETA BOUND TO THE IBB DOMAIN OF IMPORTIN ALPHA (II CRYSTAL FORM, GROWN AT LOW PH) | | Descriptor: | PROTEIN (IMPORTIN ALPHA-2 SUBUNIT), PROTEIN (IMPORTIN BETA SUBUNIT) | | Authors: | Cingolani, G, Petosa, C, Weis, K, Muller, C.W. | | Deposit date: | 1999-05-04 | | Release date: | 1999-05-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of importin-beta bound to the IBB domain of importin-alpha.
Nature, 399, 1999
|
|
1QGK
 
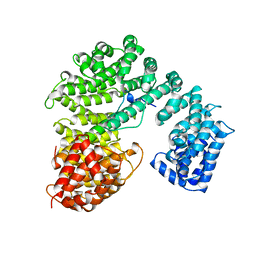 | | STRUCTURE OF IMPORTIN BETA BOUND TO THE IBB DOMAIN OF IMPORTIN ALPHA | | Descriptor: | PROTEIN (IMPORTIN ALPHA-2 SUBUNIT), PROTEIN (IMPORTIN BETA SUBUNIT) | | Authors: | Cingolani, G, Petosa, C, Weis, K, Muller, C.W. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of importin-beta bound to the IBB domain of importin-alpha.
Nature, 399, 1999
|
|
1H2O
 
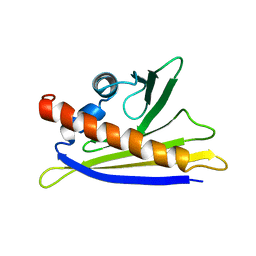 | | SOLUTION STRUCTURE OF THE MAJOR CHERRY ALLERGEN PRU AV 1 MUTANT E45W | | Descriptor: | MAJOR ALLERGEN PRU AV 1 | | Authors: | Neudecker, P, Lehmann, K, Nerkamp, J, Schweimer, K, Sticht, H, Boehm, M, Scheurer, S, Vieths, S, Roesch, P. | | Deposit date: | 2002-08-12 | | Release date: | 2003-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mutational Epitope Analysis of Pru Av 1 and Api G 1, the Major Allergens of Cherry (Prunus Avium) and Celery (Apium Graveolens): Correlating Ige Reactivity with Three-Dimensional Structure
Biochem.J., 376, 2003
|
|
6UFE
 
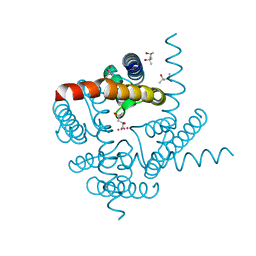 | | The structure of a potassium selective ion channel at atomic resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Transporter | | Authors: | Langan, P.S, Vandavasi, V.G, Sullivan, B, Afonine, P.V, Weiss, K.L. | | Deposit date: | 2019-09-24 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The structure of a potassium-selective ion channel reveals a hydrophobic gate regulating ion permeation.
Iucrj, 7, 2020
|
|
1GS9
 
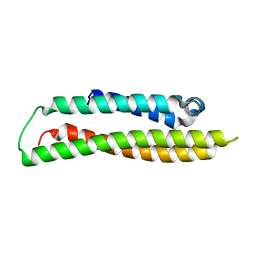 | | Apolipoprotein E4, 22k domain | | Descriptor: | APOLIPOPROTEIN E | | Authors: | Verderame, J.R, Kantardjieff, K, Segelke, B, Weisgraber, K, Rupp, B. | | Deposit date: | 2002-01-02 | | Release date: | 2003-06-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the 22K Domain of Human Apolipoprotein E4
To be Published
|
|
6TF4
 
 | |
6ZL9
 
 | |
8ODW
 
 | | Crystal structure of LbmA Ox-ACP didomain in complex with NADP and ethyl glycinate from the lobatamide PKS (Gynuella sunshinyii) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Francois, R.M.M, Fraley, A.E, Piel, J, Weissman, K.J, Gruez, A. | | Deposit date: | 2023-03-09 | | Release date: | 2023-05-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Modular Oxime Formation by a trans-AT Polyketide Synthase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6ER0
 
 | | 6th KOW domain of human hSpt5 | | Descriptor: | Transcription elongation factor SPT5 | | Authors: | Hahn, L, Schweimer, K, Roesch, P, Woehrl, B.M. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and nucleic acid binding properties of KOW domains 4 and 6-7 of human transcription elongation factor DSIF.
Sci Rep, 8, 2018
|
|
6BIT
 
 | | SIRPalpha antibody complex | | Descriptor: | KWAR23 Fab heavy chain, KWAR23 Fab light chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | Authors: | Ring, N.G, Herndler-Brandstetter, D, Weiskopf, K, Shan, L, Volkmer, J.P, George, B.M, Lietzenmayer, M, McKenna, K.M, Naik, T.J, McCarty, A, Zheng, Y, Ring, A.M, Flavell, R.A, Weissman, I.L. | | Deposit date: | 2017-11-03 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Anti-SIRP alpha antibody immunotherapy enhances neutrophil and macrophage antitumor activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
