7N0J
 
 | | Structure of YebY from E. coli K12 | | Descriptor: | YebY | | Authors: | Hadley, R.C, Rosenzweig, A.C. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-09 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The copper-linked Escherichia coli AZY operon: Structure, metal binding, and a possible physiological role in copper delivery.
J.Biol.Chem., 298, 2022
|
|
5K67
 
 | | Designed Artificial Cupredoxins | | Descriptor: | GLYCEROL, Streptavidin, [CuII(biot-pr-dpea)]2+ | | Authors: | Mann, S.I, Heinisch, T, Weitz, A.C, Hendrich, M.R, Ward, T.R, Borovik, A.S. | | Deposit date: | 2016-05-24 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Modular Artificial Cupredoxins.
J.Am.Chem.Soc., 138, 2016
|
|
6SBJ
 
 | | X-ray structure of mus musculus Fumarylacetoacetate hydrolase domain containing protein 1 (FAHD1) apo-form uuncomplexed | | Descriptor: | Acylpyruvase FAHD1, mitochondrial, CHLORIDE ION, ... | | Authors: | Rupp, B, Naschberger, A, Weiss, A.K.H. | | Deposit date: | 2019-07-21 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and functional comparison of fumarylacetoacetate domain containing protein 1 in human and mouse.
Biosci.Rep., 40, 2020
|
|
6SBI
 
 | | X-ray structure of murine Fumarylacetoacetate hydrolase domain containing protein 1 (FAHD1) in complex with inhibitor oxalate | | Descriptor: | Acylpyruvase FAHD1, mitochondrial, CHLORIDE ION, ... | | Authors: | Rupp, B, Naschberger, A, Weiss, A.K.H. | | Deposit date: | 2019-07-21 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional comparison of fumarylacetoacetate domain containing protein 1 in human and mouse.
Biosci.Rep., 40, 2020
|
|
6W25
 
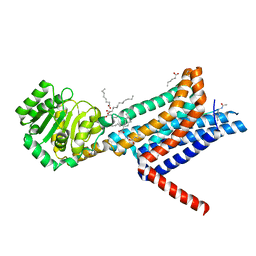 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with SHU9119 | | Descriptor: | CALCIUM ION, Melanocortin receptor 4,GlgA glycogen synthase,Melanocortin receptor 4, OLEIC ACID, ... | | Authors: | Yu, J, Gimenez, L.E, Hernandez, C.C, Wu, Y, Wein, A.H, Han, G.W, McClary, K, Mittal, S.R, Burdsall, K, Stauch, B, Wu, L, Stevens, S.N, Peisley, A, Williams, S.Y, Chen, V, Millhauser, G.L, Zhao, S, Cone, R.D, Stevens, R.C. | | Deposit date: | 2020-03-04 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Determination of the melanocortin-4 receptor structure identifies Ca2+as a cofactor for ligand binding.
Science, 368, 2020
|
|
6FOG
 
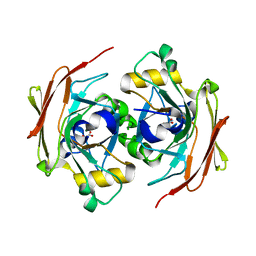 | |
6FOH
 
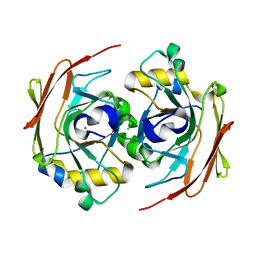 | |
5W5G
 
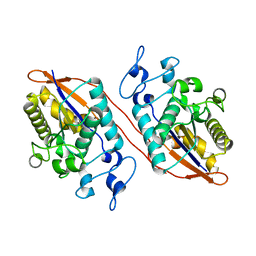 | | Structure of Human Sts-1 histidine phosphatase domain | | Descriptor: | Ubiquitin-associated and SH3 domain-containing protein B | | Authors: | Zhou, W, Yin, Y, Weinheimer, A.W, Kaur, N, Carpino, N, French, J.B. | | Deposit date: | 2017-06-15 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural and Functional Characterization of the Histidine Phosphatase Domains of Human Sts-1 and Sts-2.
Biochemistry, 56, 2017
|
|
8SQW
 
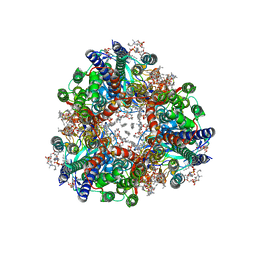 | | particulate methane monooxygenase crosslinked with 2,2,2-trifluoroethanol bound | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Tucci, F.J, Rosenzweig, A.C. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.16 Å) | | Cite: | Product analog binding identifies the copper active site of particulate methane monooxygenase.
Nat Catal, 6, 2023
|
|
8SR1
 
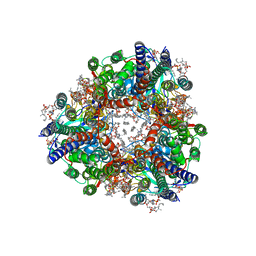 | | particulate methane monooxygenase crosslinked with 4,4,4-trifluorobutanol bound | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, 4,4,4-trifluorobutan-1-ol, ... | | Authors: | Tucci, F.J, Rosenzweig, A.C. | | Deposit date: | 2023-05-05 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.18 Å) | | Cite: | Product analog binding identifies the copper active site of particulate methane monooxygenase.
Nat Catal, 6, 2023
|
|
6RI7
 
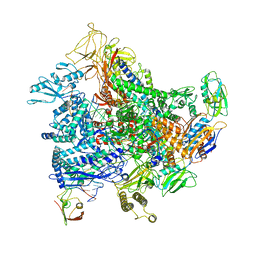 | | Cryo-EM structure of E. coli RNA polymerase elongation complex bound to GreB transcription factor | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | Deposit date: | 2019-04-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
8OL6
 
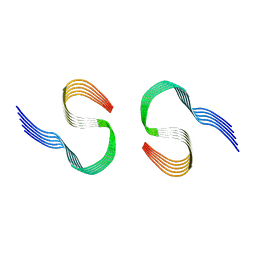 | | Murine type II Abeta fibril from tgAPPSwe mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
8OLN
 
 | | DI1 Abeta fibril from tg-SwDI mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
8OLQ
 
 | | DI3 Abeta fibril from tg-SwDI mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
8OL2
 
 | | Murine type II Abeta fibril from APP23 mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
8OL3
 
 | | Murine type III Abeta fibril from APP/PS1 mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
8OL5
 
 | | Murine type II Abeta fibril from ARTE10 mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
8OL7
 
 | | MurineArc type I Abeta fibril from tg-APPArcSwe mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
8OLG
 
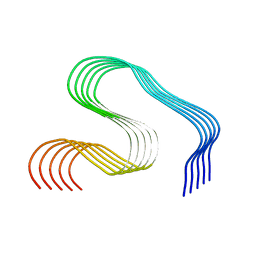 | | DI2 Abeta fibril from tg-SwDI mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
8OLO
 
 | | Murine type III Abeta fibril from ARTE10 mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
6RIP
 
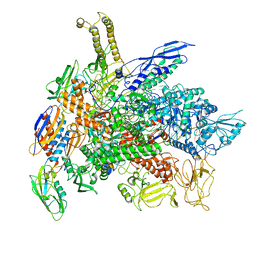 | | Cryo-EM structure of E. coli RNA polymerase backtracked elongation complex in swiveled state | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | Deposit date: | 2019-04-24 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
6RI9
 
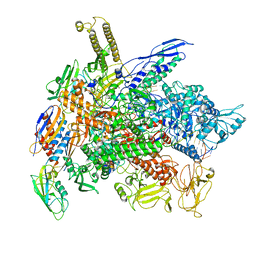 | | Cryo-EM structure of E. coli RNA polymerase backtracked elongation complex in non-swiveled state | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | Deposit date: | 2019-04-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
6RIN
 
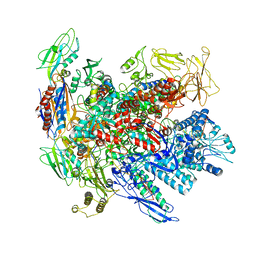 | | Cryo-EM structure of E. coli RNA polymerase backtracked elongation complex bound to GreB transcription factor | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | Deposit date: | 2019-04-24 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
6RH3
 
 | | Cryo-EM structure of E. coli RNA polymerase elongation complex bound to CTP substrate | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | Deposit date: | 2019-04-18 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
6E1C
 
 | |
