2AOH
 
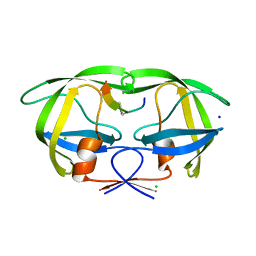 | | Crystal structure analysis of HIV-1 Protease mutant V82A with a substrate analog P6-PR | | 分子名称: | CHLORIDE ION, PEPTIDE INHIBITOR, POL POLYPROTEIN, ... | | 著者 | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2005-08-12 | | 公開日 | 2006-01-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2AVS
 
 | | kinetics, stability, and structural changes in high resolution crystal structures of HIV-1 protease with drug resistant mutations L24I, I50V, and G73S | | 分子名称: | ACETIC ACID, DIMETHYL SULFOXIDE, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, ... | | 著者 | Liu, F, Boross, P.I, Wang, Y.F, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | 登録日 | 2005-08-30 | | 公開日 | 2006-01-24 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Kinetic, stability, and structural changes in high-resolution crystal structures of HIV-1 protease with drug-resistant mutations L24I, I50V, and G73S.
J.Mol.Biol., 354, 2005
|
|
3UCB
 
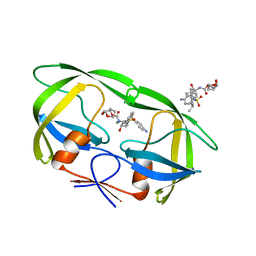 | | Crystal Structure of Multidrug Resistant HIV-1 Protease Clinical Isolate PR20 in Complex with Darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | 著者 | Agniswamy, J, Chen-Hsiang, S, Aniana, A, Sayer, J.M, Louis, J.M, Weber, I.T. | | 登録日 | 2011-10-26 | | 公開日 | 2012-03-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | HIV-1 protease with 20 mutations exhibits extreme resistance to clinical inhibitors through coordinated structural rearrangements.
Biochemistry, 51, 2012
|
|
3UFN
 
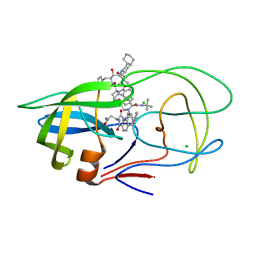 | | Crystal Structure of Multidrug Resistant HIV-1 Protease Clinical Isolate PR20 in Complex with Saquinavir | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, HIV-1 protease | | 著者 | Agniswamy, J, Chen-Hsiang, S, Aniana, A, Sayer, J.M, Louis, J.M, Weber, I.T. | | 登録日 | 2011-11-01 | | 公開日 | 2012-03-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | HIV-1 protease with 20 mutations exhibits extreme resistance to clinical inhibitors through coordinated structural rearrangements.
Biochemistry, 51, 2012
|
|
3NU3
 
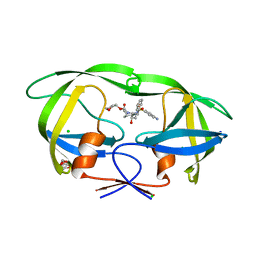 | | Wild Type HIV-1 Protease with Antiviral Drug Amprenavir | | 分子名称: | CHLORIDE ION, GLYCEROL, Protease, ... | | 著者 | Wang, Y.-F, Kovalevsky, A.Y, Weber, I.T. | | 登録日 | 2010-07-06 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.02 Å) | | 主引用文献 | Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters.
Febs J., 277, 2010
|
|
3NYC
 
 | |
3NU6
 
 | |
3NU5
 
 | | Crystal Structure of HIV-1 Protease Mutant I50V with Antiviral Drug Amprenavir | | 分子名称: | ACETATE ION, CHLORIDE ION, SODIUM ION, ... | | 著者 | Wang, Y.-F, Shen, C.H, Weber, I.T. | | 登録日 | 2010-07-06 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters.
Febs J., 277, 2010
|
|
3NYF
 
 | |
3NNE
 
 | | Crystal structure of choline oxidase S101A mutant | | 分子名称: | ACETATE ION, Choline oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Wang, Y.-F, Finnegan, S, Yuan, H, Orville, A.M, Weber, I.T, Gadda, G. | | 登録日 | 2010-06-23 | | 公開日 | 2010-07-28 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Structural and kinetic studies on the Ser101Ala variant of choline oxidase: Catalysis by compromise.
Arch.Biochem.Biophys., 501, 2010
|
|
3NUJ
 
 | |
3NYE
 
 | |
3NU4
 
 | | Crystal Structure of HIV-1 Protease Mutant V32I with Antiviral Drug Amprenavir | | 分子名称: | CHLORIDE ION, SODIUM ION, protease, ... | | 著者 | Wang, Y.-F, Kovalevsky, A.Y, Weber, I.T. | | 登録日 | 2010-07-06 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters.
Febs J., 277, 2010
|
|
3NUO
 
 | |
3NU9
 
 | |
3MWS
 
 | | Crystal Structure of Group N HIV-1 Protease | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, HIV-1 Protease | | 著者 | Sayer, J.M, Agniswamy, J, Weber, I.T, Louis, J.M. | | 登録日 | 2010-05-06 | | 公開日 | 2011-03-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.09 Å) | | 主引用文献 | Autocatalytic maturation, physical/chemical properties, and crystal structure of group N HIV-1 protease: relevance to drug resistance.
Protein Sci., 19, 2010
|
|
3PWM
 
 | | HIV-1 Protease Mutant L76V with Darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | 著者 | Zhang, Y, Weber, I.T. | | 登録日 | 2010-12-08 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | The L76V Drug Resistance Mutation Decreases the Dimer Stability and Rate of Autoprocessing of HIV-1 Protease by Reducing Internal Hydrophobic Contacts.
Biochemistry, 50, 2011
|
|
3OK9
 
 | | Crystal structure of wild-type HIV-1 protease with new oxatricyclic designed inhibitor GRL-0519A | | 分子名称: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, GLYCEROL, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2010-08-24 | | 公開日 | 2010-09-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | Probing Multidrug-Resistance and Protein-Ligand Interactions with Oxatricyclic Designed Ligands in HIV-1 Protease Inhibitors.
Chemmedchem, 5, 2010
|
|
3PWR
 
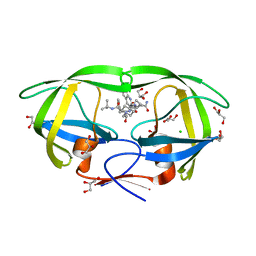 | | HIV-1 Protease Mutant L76V complexed with Saquinavir | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, GLYCEROL, ... | | 著者 | Zhang, Y, Weber, I.T. | | 登録日 | 2010-12-08 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The L76V Drug Resistance Mutation Decreases the Dimer Stability and Rate of Autoprocessing of HIV-1 Protease by Reducing Internal Hydrophobic Contacts.
Biochemistry, 50, 2011
|
|
2NNP
 
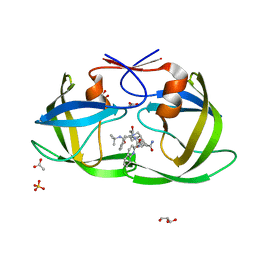 | | Crystal structure analysis of HIV-1 protease mutant I84V with a inhibitor saquinavir | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, ACETIC ACID, GLYCEROL, ... | | 著者 | Tie, Y, Kovalevsky, A.Y, Boross, P, Wang, Y.F, Ghosh, A.K, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2006-10-24 | | 公開日 | 2007-03-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir.
Proteins, 67, 2007
|
|
2NMZ
 
 | | Crystal structure analysis of HIV-1 protease mutant V82A with a inhibitor saquinavir | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, PROTEASE, SULFATE ION | | 著者 | Tie, Y, Kovalevsky, A.Y, Boross, P, Wang, Y.F, Ghosh, A.K, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2006-10-23 | | 公開日 | 2007-03-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (0.97 Å) | | 主引用文献 | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir.
Proteins, 67, 2007
|
|
2NNK
 
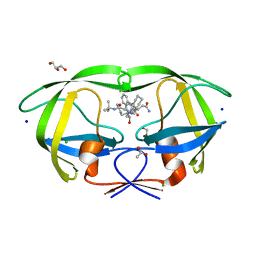 | | Crystal structure analysis of HIV-1 protease mutant I84V with a inhibitor saquinavir | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, ACETIC ACID, CHLORIDE ION, ... | | 著者 | Tie, Y, Kovalevsky, A.Y, Boross, P, Wang, Y.F, Ghosh, A.K, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2006-10-24 | | 公開日 | 2007-03-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir.
Proteins, 67, 2007
|
|
2NMY
 
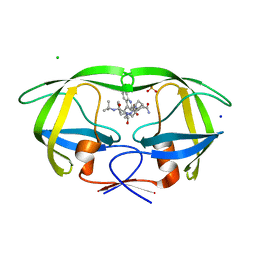 | | Crystal structure analysis of HIV-1 protease mutant V82A with a inhibitor saquinavir | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, PROTEASE, ... | | 著者 | Tie, Y, Kovalevsky, A.Y, Boross, P, Wang, Y.F, Ghosh, A.K, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2006-10-23 | | 公開日 | 2007-03-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir.
Proteins, 67, 2007
|
|
2OGT
 
 | | Crystal Structure of the Geobacillus Stearothermophilus Carboxylesterase EST55 at pH 6.8 | | 分子名称: | GLYCEROL, IODIDE ION, Thermostable carboxylesterase Est50 | | 著者 | Liu, P, Ewis, H.E, Tai, P.C, Lu, C.D, Weber, I.T. | | 登録日 | 2007-01-08 | | 公開日 | 2007-02-13 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Crystal Structure of the Geobacillus stearothermophilus Carboxylesterase Est55 and Its Activation of Prodrug CPT-11.
J.Mol.Biol., 367, 2007
|
|
2OGS
 
 | | Crystal Structure of the GEOBACILLUS STEAROTHERMOPHILUS Carboxylesterase EST55 at pH 6.2 | | 分子名称: | IODIDE ION, Thermostable carboxylesterase Est50 | | 著者 | Liu, P, Ewis, H.E, Tai, P.C, Lu, C.D, Weber, I.T. | | 登録日 | 2007-01-08 | | 公開日 | 2007-02-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Crystal Structure of the Geobacillus stearothermophilus Carboxylesterase Est55 and Its Activation of Prodrug CPT-11.
J.Mol.Biol., 367, 2007
|
|
