5QH8
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with PCM-0102558 | | Descriptor: | 1-[(2S)-2-(4-methoxyphenyl)-2,3,6,7-tetrahydro-1H-azepin-1-yl]ethan-1-one, ACETATE ION, Peroxisomal coenzyme A diphosphatase NUDT7 | | Authors: | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2018-05-15 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
1J85
 
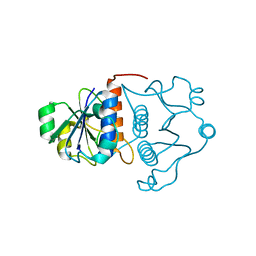 | | Structure of YibK from Haemophilus influenzae (HI0766), a truncated sequence homolog of tRNA (guanosine-2'-O-) methyltransferase (SpoU) | | Descriptor: | YibK | | Authors: | Lim, K, Zhang, H, Toedt, J, Tempcyzk, A, Krajewski, W, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-20 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the YibK methyltransferase from Haemophilus influenzae
(HI0766): A cofactor bound at a site formed by a knot
Proteins, 51, 2003
|
|
2GUH
 
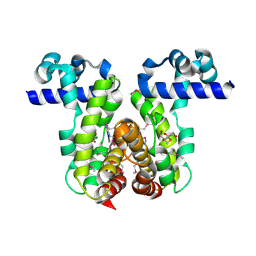 | | Crystal Structure of the Putative TetR-family Transcriptional Regulator from Rhodococcus sp. RHA1 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Putative TetR-family transcriptional regulator | | Authors: | Kim, Y, Joachimiak, A, Skarina, T, Zheng, H, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-04-30 | | Release date: | 2006-05-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of the Putative TetR-family Transcriptional Regulator from Rhodococcus sp. RHA1
To be Published
|
|
2C2N
 
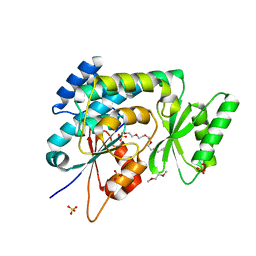 | | Structure of human mitochondrial malonyltransferase | | Descriptor: | 1,2-DIMETHOXYETHANE, 2-(2-ETHOXYETHOXY)ETHANOL, 3,6,9,12,15-PENTAOXAHEPTADECAN-1-OL, ... | | Authors: | Wu, X, Bunkoczi, G, Smee, C, Arrowsmith, C, Sundstrom, M, Weigelt, J, Edwards, A, von Delft, F, Oppermann, U. | | Deposit date: | 2005-09-29 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for Different Specificities of Acyltransferases Associated with the Human Cytosolic and Mitochondrial Fatty Acid Synthases.
Chem.Biol., 16, 2009
|
|
2H0U
 
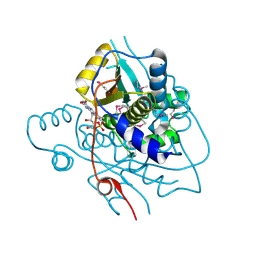 | | Crystal structure of NAD(P)H-flavin oxidoreductase from Helicobacter pylori | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH-flavin oxidoreductase | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-05-15 | | Release date: | 2006-06-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of NAD(P)H-flavin oxidoreductase from Helicobacter pylori
To be Published
|
|
1RXX
 
 | | Structure of arginine deiminase | | Descriptor: | Arginine deiminase | | Authors: | Galkin, A, Kulakova, L, Sarikaya, E, Lim, K, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insight into arginine degradation by arginine deiminase, an antibacterial and parasite drug target.
J.Biol.Chem., 279, 2004
|
|
3OS6
 
 | | Crystal structure of putative 2,3-dihydroxybenzoate-specific isochorismate synthase, DhbC from Bacillus anthracis. | | Descriptor: | GLYCEROL, Isochorismate synthase DhbC, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Domagalski, M.J, Chruszcz, M, Skarina, T, Onopriyenko, O, Cymborowski, M, Savchenko, A, Edwards, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-08 | | Release date: | 2010-10-20 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of isochorismate synthase DhbC from Bacillus anthracis.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
2BVA
 
 | | Crystal structure of the human P21-activated kinase 4 | | Descriptor: | P21-ACTIVATED KINASE 4 | | Authors: | Debreczeni, J.E, Bunkoczi, G, Eswaran, J, Filippakopoulos, P, Das, S, Fedorov, O, Sundstrom, M, Arrowsmith, C, Edwards, A, von Delft, F, Knapp, S. | | Deposit date: | 2005-06-23 | | Release date: | 2005-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the P21-Activated Kinases Pak4, Pak5, and Pak6 Reveal Catalytic Domain Plasticity of Active Group II Paks.
Structure, 15, 2007
|
|
2H8O
 
 | | The 1.6A crystal structure of the geranyltransferase from Agrobacterium tumefaciens | | Descriptor: | Geranyltranstransferase | | Authors: | Zhang, R, Xu, X, Gu, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-06-07 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6A crystal structure of the geranyltransferase from Agrobacterium tumefaciens
To be Published
|
|
2A3K
 
 | | Crystal Structure of the Human Protein Tyrosine Phosphatase, PTPN7 (HePTP, Hematopoietic Protein Tyrosine Phosphatase) | | Descriptor: | PHOSPHATE ION, protein tyrosine phosphatase, non-receptor type 7, ... | | Authors: | Barr, A, Turnbull, A.P, Das, S, Eswaran, J, Debreczeni, J.E, Longmann, E, Smee, C, Burgess, N, Gileadi, O, Sundstrom, M, Arrowsmith, C, Edwards, A, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-24 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of human receptor protein tyrosine phosphatase kappa phosphatase domain 1.
Protein Sci., 15, 2006
|
|
1SDI
 
 | | 1.65 A structure of Escherichia coli ycfC gene product | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, Hypothetical protein ycfC | | Authors: | Borek, D, Otwinowski, Z, Chen, Y, Skarina, T, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-13 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural analysis of Escherichia coli ycfC gene product
To be Published
|
|
2HHG
 
 | | Structure of Protein of Unknown Function RPA3614, Possible Tyrosine Phosphatase, from Rhodopseudomonas palustris CGA009 | | Descriptor: | Hypothetical protein RPA3614, PHOSPHATE ION, SODIUM ION | | Authors: | Binkowski, T.A, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-06-28 | | Release date: | 2006-07-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Hypothetical protein RPA3614 from Rhodopseudomonas palustris CGA009
To be published
|
|
2HJS
 
 | | The structure of a probable aspartate-semialdehyde dehydrogenase from Pseudomonas aeruginosa | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, USG-1 protein homolog | | Authors: | Cuff, M.E, Evdokimova, E, Kudritska, M, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-06-30 | | Release date: | 2006-08-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a probable aspartate-semialdehyde dehydrogenase from Pseudomonas aeruginosa
To be published
|
|
1T57
 
 | | Crystal Structure of the Conserved Protein MTH1675 from Methanobacterium thermoautotrophicum | | Descriptor: | Conserved Protein MTH1675, FLAVIN MONONUCLEOTIDE, MAGNESIUM ION | | Authors: | Kim, Y, Joachimiak, A, Saridakis, V, Xu, X, Arrowsmith, C.H, Christendat, D, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-03 | | Release date: | 2004-08-03 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Conserved Protein MTH1675 from Methanobacterium thermoautotrophicum
To be Published
|
|
2HRZ
 
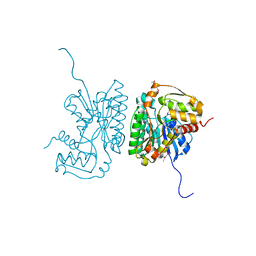 | | The crystal structure of the nucleoside-diphosphate-sugar epimerase from Agrobacterium tumefaciens | | Descriptor: | Nucleoside-diphosphate-sugar epimerase | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-07-20 | | Release date: | 2006-08-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of the nucleoside-diphosphate-sugar epimerase from Agrobacterium tumefaciens
To be Published
|
|
2HKU
 
 | | Structural Genomics, the crystal structure of a putative transcriptional regulator from Rhodococcus sp. RHA1 | | Descriptor: | 1,2-ETHANEDIOL, A putative transcriptional regulator, TETRAETHYLENE GLYCOL | | Authors: | Tan, K, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-07-05 | | Release date: | 2006-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a transcriptional regulator from Rhodococcus sp. RHA1
To be Published
|
|
2I71
 
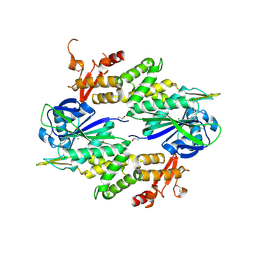 | | Crystal structure of a Conserved Protein of Unknown Function from Sulfolobus solfataricus P2 | | Descriptor: | Hypothetical protein, MAGNESIUM ION | | Authors: | Tan, K, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-30 | | Release date: | 2006-10-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a Conserved hypothetical protein from Sulfolobus solfataricus P2
To be Published
|
|
2HXI
 
 | | Structural Genomics, the crystal structure of a putative transcriptional regulator from Streptomyces coelicolor A3(2) | | Descriptor: | Putative transcriptional regulator | | Authors: | Tan, K, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-03 | | Release date: | 2006-09-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a putative transcriptional regulator TetR from Streptomyces coelicolor A3(2)
To be Published
|
|
2OWI
 
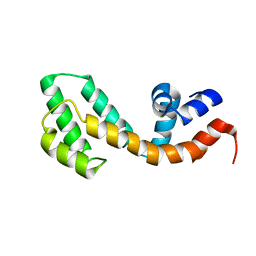 | | Solution structure of the RGS domain from human RGS18 | | Descriptor: | Regulator of G-protein signaling 18 | | Authors: | Higman, V.A, Leidert, M, Bray, J, Elkins, J, Soundararajan, M, Doyle, D.A, Gileadi, C, Phillips, C, Schoch, G, Yang, X, Brockmann, C, Schmieder, P, Diehl, A, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, Oschkinat, H, Ball, L.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-16 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4BKN
 
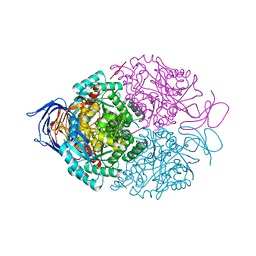 | | Human Dihydropyrimidinase-related protein 3 (DPYSL3) | | Descriptor: | DIHYDROPYRIMIDINASE-RELATED PROTEIN 3 | | Authors: | Mathea, S, Elkins, J.M, Alegre-Abarrategui, J, Shrestha, L, Burgess-Brown, N, Puranik, S, Coutandin, D, Bradley, A, Vollmar, M, von Delft, F, Bountra, C, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2013-04-26 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Dpysl3
To be Published
|
|
5L78
 
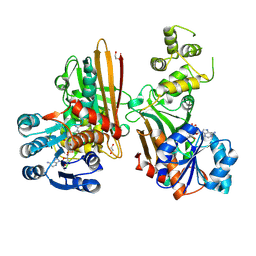 | | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain (in NAD+ bound form) | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde synthase, mitochondrial, ... | | Authors: | Kopec, J, Pena, I.A, Rembeza, E, Strain-Damerell, C, Chalk, R, Borkowska, O, Goubin, S, Velupillai, S, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, Arruda, P, Yue, W.W. | | Deposit date: | 2016-06-02 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain (in NAD+ bound form)
To Be Published
|
|
5RLJ
 
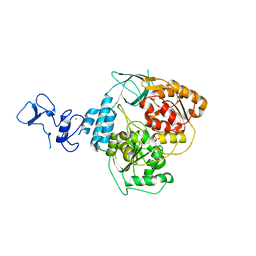 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z1407673036 | | Descriptor: | (2S)-2-phenylpropane-1-sulfonamide, Helicase, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.879 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RM2
 
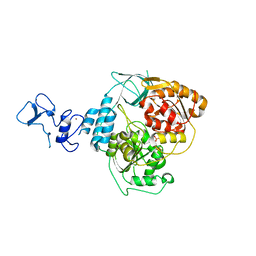 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z1741964527 | | Descriptor: | 1-(diphenylmethyl)azetidin-3-ol, Helicase, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RMH
 
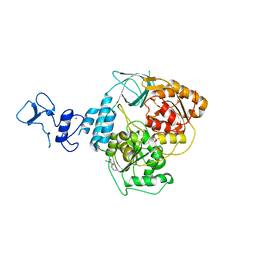 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z1101755952 | | Descriptor: | Helicase, PHOSPHATE ION, ZINC ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.021 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RL8
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z53825177 | | Descriptor: | Helicase, N-(2-fluorophenyl)ethanesulfonamide, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
