6CEV
 
 | | MBD3 MBD in complex with methylated, non-palindromic CpG DNA: alternative interpretation of crystallographic data | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*CP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*GP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain protein 3, ... | | Authors: | Liu, K, Tempel, W, Wernimont, A.K, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural analyses reveal that MBD3 is a methylated CG binder.
Febs J., 286, 2019
|
|
6CEU
 
 | | MBD3 MBD in complex with methylated, non-palindromic CpG DNA: alternative interpretation of crystallographic data | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*CP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*GP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain protein 3, ... | | Authors: | Liu, K, Tempel, W, Wernimont, A.K, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural analyses reveal that MBD3 is a methylated CG binder.
Febs J., 286, 2019
|
|
6CC8
 
 | | Crystal structure MBD3 MBD domain in complex with methylated CpG DNA | | Descriptor: | Methyl-CpG-binding domain protein 3, UNKNOWN ATOM OR ION, methylated CpG DNA | | Authors: | Liu, K, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-06 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analyses reveal that MBD3 is a methylated CG binder.
Febs J., 286, 2019
|
|
6CE8
 
 | | Crystal structure of fragment 2-(Benzo[d]thiazol-2-yl)acetic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | (1,3-benzothiazol-2-yl)acetic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Ravichandran, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
6CEC
 
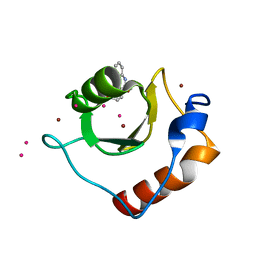 | | Crystal structure of fragment 3-(3-Methoxy-2-quinoxalinyl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(3-methoxyquinoxalin-2-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Franzoni, I, Ravichandran, M, Lautens, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
3E7Q
 
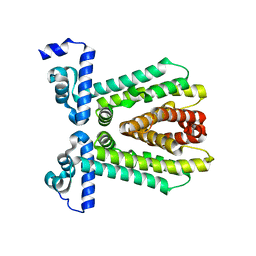 | | The crystal structure of the putative transcriptional regulator from Pseudomonas aeruginosa PAO1 | | Descriptor: | transcriptional regulator | | Authors: | Zhang, R, Skarina, T, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-18 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the putative transcriptional regulator from Pseudomonas aeruginosa PAO1
To be Published
|
|
6CCG
 
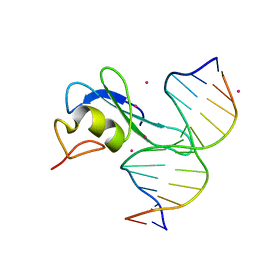 | | Crystal structure MBD3 MBD domain in complex with methylated CpG DNA | | Descriptor: | DNA, Methyl-CpG-binding domain protein 3, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analyses reveal that MBD3 is a methylated CG binder.
Febs J., 286, 2019
|
|
3EC8
 
 | | The crystal structure of the RA domain of FLJ10324 (RADIL) | | Descriptor: | CHLORIDE ION, GLYCEROL, LEAD (II) ION, ... | | Authors: | Wisniewska, M, Lehtio, L, Andersson, J, Arrowsmith, C.H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Schueler, H, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Welin, M, Wikstrom, M, Berglund, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-29 | | Release date: | 2008-09-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of the RA domain of FLJ10324 (RADIL)
to be published
|
|
2X47
 
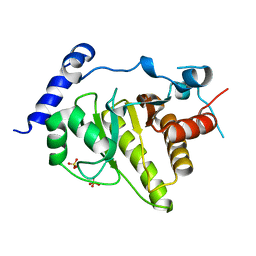 | | Crystal structure of human MACROD1 | | Descriptor: | MACRO DOMAIN-CONTAINING PROTEIN 1, SULFATE ION | | Authors: | Vollmar, M, Phillips, C, Mehrotra, P.V, Ahel, I, Krojer, T, Yue, W, Ugochukwu, E, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Gileadi, O. | | Deposit date: | 2010-01-28 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of Macro Domain Proteins as Novel O-Acetyl-Adp-Ribose Deacetylases.
J.Biol.Chem., 286, 2011
|
|
3CBQ
 
 | | Crystal structure of the human REM2 GTPase with bound GDP | | Descriptor: | 1,2-ETHANEDIOL, GTP-binding protein REM 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nedyalkova, L, Shen, Y, Tong, Y, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-22 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of the human REM2 GTPase with bound GDP.
To be Published
|
|
3C9H
 
 | | Crystal structure of the substrate binding protein of the ABC transporter from Agrobacterium tumefaciens | | Descriptor: | ABC transporter, substrate binding protein, CITRIC ACID, ... | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-15 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of the substrate binding protein of the ABC transporter from Agrobacterium tumefaciens.
To be Published
|
|
3CC9
 
 | | Crystal structure of Plasmodium vivax putative polyprenyl pyrophosphate synthase in complex with geranylgeranyl diphosphate | | Descriptor: | GERANYLGERANYL DIPHOSPHATE, Putative farnesyl pyrophosphate synthase, SODIUM ION | | Authors: | Wernimont, A.K, Dunford, J, Lew, J, Zhao, Y, Kozieradzki, I, Cossar, D, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-25 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Plasmodium vivax putative polyprenyl pyrophosphate synthase in complex with geranylgeranyl diphosphate.
To be Published
|
|
4GF1
 
 | | Crystal Structure of Certhrax | | Descriptor: | Putative ADP-ribosyltransferase Certhrax, UNKNOWN ATOM OR ION | | Authors: | Hong, B.S, Dimov, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-08-02 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Certhrax Toxin, an Anthrax-related ADP-ribosyltransferase from Bacillus cereus.
J.Biol.Chem., 287, 2012
|
|
5K5F
 
 | | NMR structure of the HLTF HIRAN domain | | Descriptor: | Helicase-like transcription factor | | Authors: | Bezsonova, I, Neculai, D, Korzhnev, D, Weigelt, J, Bountra, C, Edwards, A, Arrowsmith, C, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-05-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the HLTF HIRAN domain
To Be Published
|
|
5E1M
 
 | | Crystal structure of NTMT1 in complex with PPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-29 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
3ELK
 
 | | Crystal structure of putative transcriptional regulator TA0346 from Thermoplasma acidophilum | | Descriptor: | CHLORIDE ION, Putative transcriptional regulator TA0346 | | Authors: | Grantz Saskova, K, Chruszcz, M, Evdokimova, E, Egorova, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-22 | | Release date: | 2008-09-30 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative transcriptional regulator TA0346 from Thermoplasma acidophilum
To be Published
|
|
2FVL
 
 | | Crystal structure of human 3-alpha hydroxysteroid/dihydrodiol dehydrogenase (AKR1C4) complexed with NADP+ | | Descriptor: | Aldo-keto reductase family 1, member C4, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ugochukwu, E, Smee, C, Guo, K, Lukacik, P, Kavanagh, K, Debreczeni, J.E, von Delft, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-31 | | Release date: | 2006-02-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human 3-alpha hydroxysteroid/dihydrodiol dehydrogenase (AKR1C4) complexed with NADP+
To be Published
|
|
3Q1D
 
 | | The B-box domain of Trim54 | | Descriptor: | Tripartite motif-containing protein 54, ZINC ION | | Authors: | Walker, J.R, Yermekbayeva, L, Mackenzie, F, Dong, A, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Dhe-Paganon, S. | | Deposit date: | 2010-12-17 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The B-Box Domain of Trim54
To be Published
|
|
2G0N
 
 | | The Crystal Structure of the Human RAC3 in complex with GDP and Chloride | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ugochukwu, E, Yang, X, Zao, Y, Elkins, J, Gileadi, C, Burgess, N, Colebrook, S, Gileadi, O, Fedorov, O, Bunkoczi, G, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, von Delft, F, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-02-13 | | Release date: | 2006-05-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of the Human RAC3 in complex with GDP and Chloride
To be Published
|
|
6W6D
 
 | | Crystal Structure of Human Protein arginine N-methyltransferase 6 (PRMT6) in complex with SGC6870 inhibitor | | Descriptor: | (5R)-4-(5-bromothiophene-2-carbonyl)-5-(3,5-dimethylphenyl)-7-methyl-1,3,4,5-tetrahydro-2H-1,4-benzodiazepin-2-one, Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Halabelian, L, Zeng, H, Dong, A, Jin, J, Shen, Y, Kaniskan, H.U, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A First-in-Class, Highly Selective and Cell-Active Allosteric Inhibitor of Protein Arginine Methyltransferase 6.
J.Med.Chem., 64, 2021
|
|
3CKK
 
 | | Crystal structure of human methyltransferase-like protein 1 | | Descriptor: | GLYCEROL, S-ADENOSYLMETHIONINE, tRNA (guanine-N(7)-)-methyltransferase | | Authors: | Dong, A, Zeng, H, Dobrovetsky, E, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Plotnikov, A.N, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-16 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of human methyltransferase-like protein 1.
To be Published
|
|
3CNV
 
 | | Crystal structure of the ligand-binding domain of a putative GntR-family transcriptional regulator from Bordetella bronchiseptica | | Descriptor: | CHLORIDE ION, CITRATE ANION, Putative GntR-family transcriptional regulator | | Authors: | Zimmerman, M.D, Xu, X, Cui, H, Filippova, E.V, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-26 | | Release date: | 2008-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the ligand-binding domain of a putative GntR-family transcriptional regulator from Bordetella bronchiseptica.
To be Published
|
|
3CXG
 
 | | Crystal structure of Plasmodium falciparum thioredoxin, PFI0790w | | Descriptor: | GLYCEROL, Putative thioredoxin, SULFATE ION | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Cossar, D, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Wilkstrom, M, Edwards, A.M, Hui, R, Hills, T, Pizarro, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-24 | | Release date: | 2008-07-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Plasmodium falciparum thioredoxin, PFI0790w.
To be Published
|
|
1YLA
 
 | | Ubiquitin-conjugating enzyme E2-25 kDa (Huntington interacting protein 2) | | Descriptor: | Ubiquitin-conjugating enzyme E2-25 kDa | | Authors: | Choe, J, Avvakumov, G.V, Newman, E.M, Mackenzie, F, Kozieradzki, I, Bochkarev, A, Sundstrom, M, Arrowsmith, C, Edwards, A, Dhe-paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of E2-25K/UBB+1 interaction leading to proteasome inhibition and neurotoxicity
J.Biol.Chem., 285, 2010
|
|
3MA8
 
 | | Crystal structure of CGD1_2040, a pyruvate kinase from cryptosporidium Parvum | | Descriptor: | CITRIC ACID, Pyruvate kinase, SULFATE ION | | Authors: | Wernimont, A.K, Hutchinson, A, Hassanali, A, Kozieradzki, I, Cossar, D, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Hills, T, Pizarro, J.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-23 | | Release date: | 2010-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of CGD1_2040, a pyruvate kinase from cryptosporidium Parvum
To be Published
|
|
