6O9H
 
 | | Mouse ECD with Fab1 | | Descriptor: | Gastric inhibitory polypeptide receptor, Heavy chain, Light chain, ... | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-22 | | Last modified: | 2020-04-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism of an antagonistic antibody against glucose-dependent insulinotropic polypeptide receptor.
Mabs, 12, 2020
|
|
3JAV
 
 | | Structure of full-length IP3R1 channel in the apo-state determined by single particle cryo-EM | | Descriptor: | Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Fan, G, Baker, M.L, Wang, Z, Baker, M.R, Sinyagovskiy, P.A, Chiu, W, Ludtke, S.J, Serysheva, I.I. | | Deposit date: | 2015-06-30 | | Release date: | 2015-10-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Gating machinery of InsP3R channels revealed by electron cryomicroscopy.
Nature, 527, 2015
|
|
8Q4G
 
 | | Thin filament from FIB milled relaxed left ventricular mouse myofibrils | | Descriptor: | Actin, alpha cardiac muscle 1, Tropomyosin alpha-1 chain | | Authors: | Tamborrini, D, Wang, Z, Wagner, T, Tacke, S, Stabrin, M, Grange, M, Kho, A.L, Bennet, P, Rees, M, Gautel, M, Raunser, S. | | Deposit date: | 2023-08-06 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure of the native myosin filament in the relaxed cardiac sarcomere.
Nature, 623, 2023
|
|
6O6B
 
 | | Rotavirus A-VP3 (RVA-VP3) | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Protein VP3 | | Authors: | Kumar, D, Yu, X, Prasad, V, Wang, Z. | | Deposit date: | 2019-03-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A sub-atomic resolution cryo-EM of full-length Rotavirus A-VP3 (RVA-VP3)
To Be Published
|
|
8HQS
 
 | | Cryo-EM structure of 8-subunit Smc5/6 head region | | Descriptor: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosome element 4, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Tong, C, Wang, Z, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2022-12-14 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8I4U
 
 | | Cryo-EM structure of 5-subunit Smc5/6 hinge region | | Descriptor: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | Authors: | Qian, L, Jun, Z, Xiang, Z, Wang, Z, Tong, C, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.73 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
4N4R
 
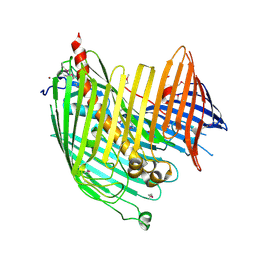 | | Structure basis of lipopolysaccharide biogenesis | | Descriptor: | CACODYLATE ION, LPS-assembly lipoprotein LptE, LPS-assembly protein LptD, ... | | Authors: | Dong, H, Xiang, Q, Wang, Z, Paterson, N.G, He, C, Zhang, Y, Wang, W, Dong, C. | | Deposit date: | 2013-10-08 | | Release date: | 2014-06-25 | | Last modified: | 2014-07-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for outer membrane lipopolysaccharide insertion.
Nature, 511, 2014
|
|
7YLM
 
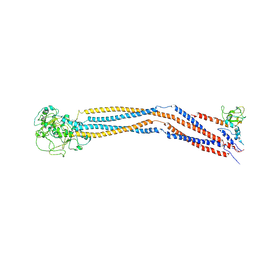 | | Cryo-EM structure of 8-subunit Smc5/6 hinge region | | Descriptor: | MMS21 isoform 1, SMC6 isoform 1, Structural maintenance of chromosomes protein 5 | | Authors: | Qian, L, Jun, Z, Xiang, Z, Wang, Z, Tong, C, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2022-07-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.17 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8I4Z
 
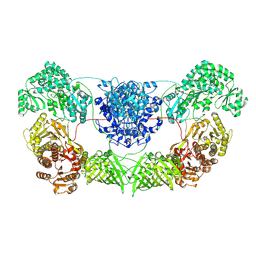 | | CalA3 with hydrolysis product | | Descriptor: | 11-oxidanylidene-11-(1~{H}-pyrrol-2-yl)undecanoic acid, Beta-ketoacyl-acyl-carrier-protein synthase I | | Authors: | Wang, J, Wang, Z. | | Deposit date: | 2023-01-21 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | C-N bond formation by a polyketide synthase.
Nat Commun, 14, 2023
|
|
8I4Y
 
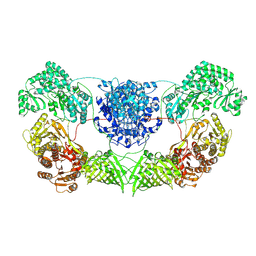 | | CalA3 complex structure with amidation product | | Descriptor: | 11-oxidanylidene-11-(1~{H}-pyrrol-2-yl)undecanoic acid, 3-HYDROXYANTHRANILIC ACID, Beta-ketoacyl-acyl-carrier-protein synthase I | | Authors: | Wang, J, Wang, Z. | | Deposit date: | 2023-01-21 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | C-N bond formation by a polyketide synthase.
Nat Commun, 14, 2023
|
|
5XE0
 
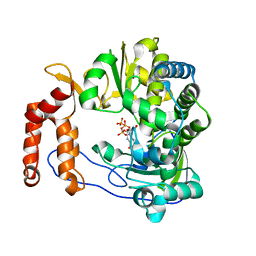 | | Crystal structure of EV-D68-3Dpol in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Genome polyprotein | | Authors: | Xie, W, Wang, C, Wang, Z, Li, Q, Wang, C. | | Deposit date: | 2017-03-30 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure and Thermostability Characterization of Enterovirus D68 3Dpol
J. Virol., 91, 2017
|
|
1J55
 
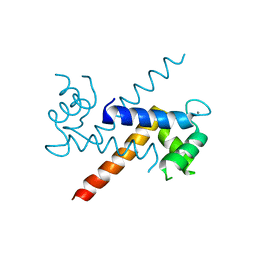 | | The Crystal Structure of Ca+-bound Human S100P Determined at 2.0A Resolution by X-ray | | Descriptor: | CALCIUM ION, S-100P PROTEIN | | Authors: | Zhang, H, Wang, G, Ding, Y, Wang, Z, Barraclough, R, Rudland, P.S, Fernig, D.G, Rao, Z. | | Deposit date: | 2002-01-25 | | Release date: | 2003-01-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure at 2A Resolution of the Ca2+-binding Protein S100P
J.Mol.Biol., 325, 2003
|
|
4CSU
 
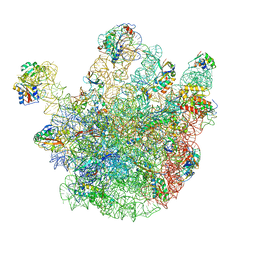 | | Cryo-EM structures of the 50S ribosome subunit bound with ObgE | | Descriptor: | 23S RRNA, 50S RIBOSOMAL PROTEIN L1, 50S RIBOSOMAL PROTEIN L11, ... | | Authors: | Feng, B, Mandava, C.S, Guo, Q, Wang, J, Cao, W, Li, N, Zhang, Y, Zhang, Y, Wang, Z, Wu, J, Sanyal, S, Lei, J, Gao, N. | | Deposit date: | 2014-03-10 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural and Functional Insights Into the Mode of Action of a Universally Conserved Obg Gtpase.
Plos Biol., 12, 2014
|
|
5KQA
 
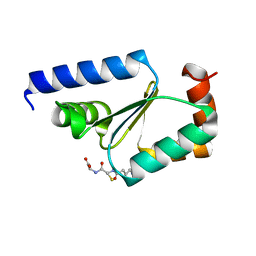 | | Crystal structure of buckwheat glutaredoxin-glutathione complex | | Descriptor: | GLUTATHIONE, Glutaredoxin-glutathione complex | | Authors: | Zhang, X, Wang, W, Zhao, Y, Wang, Z, Wang, H. | | Deposit date: | 2016-07-06 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into the binding of buckwheat glutaredoxin with GSH and regulation of its catalytic activity
J. Inorg. Biochem., 173, 2017
|
|
6L6V
 
 | | SPO1 Gp44 N-terminal region (1-55) | | Descriptor: | E3 protein | | Authors: | Liu, B, Wang, Z. | | Deposit date: | 2019-10-29 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Bacteriophage DNA Mimic Protein Employs a Non-specific Strategy to Inhibit the Bacterial RNA Polymerase.
Front Microbiol, 12, 2021
|
|
4TTH
 
 | | Crystal structure of a CDK6/Vcyclin complex with inhibitor bound | | Descriptor: | 9-cyclopentyl-N-(5-piperazin-1-ylpyridin-2-yl)pyrido[4,5]pyrrolo[1,2-d]pyrimidin-2-amine, Cyclin homolog, Cyclin-dependent kinase 6 | | Authors: | Piper, D.E, Walker, N, Wang, Z. | | Deposit date: | 2014-06-20 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of AMG 925, a FLT3 and CDK4 dual kinase inhibitor with preferential affinity for the activated state of FLT3.
J.Med.Chem., 57, 2014
|
|
6TYD
 
 | | Structure of human LDB1 in complex with SSBP2 | | Descriptor: | LIM domain-binding protein 1, Single-stranded DNA-binding protein 2 | | Authors: | Wang, H, Wang, Z, Xu, W. | | Deposit date: | 2019-08-08 | | Release date: | 2020-01-01 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Crystal structure of human LDB1 in complex with SSBP2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5J5C
 
 | | Crystal structure of ARL1-GTP and DCB domain of BIG1 complex | | Descriptor: | ADP-ribosylation factor-like protein 1, Brefeldin A-inhibited guanine nucleotide-exchange protein 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wang, R, Wang, Z, Zhang, T, Ding, J. | | Deposit date: | 2016-04-02 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for targeting BIG1 to Golgi apparatus through interaction of its DCB domain with Arl1
J Mol Cell Biol, 2016
|
|
2YJY
 
 | | A specific and modular binding code for cytosine recognition in PUF domains | | Descriptor: | 5'-R(*AP*UP*UP*GP*CP*AP*UP*AP*UP*AP)-3', PUMILIO HOMOLOG 1 | | Authors: | Dong, S, Wang, Y, Cassidy-Amstutz, C, Lu, G, Qiu, C, Bigler, R, Jezyk, M, Li, C, Hall, T.M.T, Wang, Z. | | Deposit date: | 2011-05-24 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Specific and Modular Binding Code for Cytosine Recognition in Pumilio/Fbf (Puf) RNA-Binding Domains.
J.Biol.Chem., 286, 2011
|
|
7V3L
 
 | | MERS S ectodomain trimer in complex with neutralizing antibody 6516 | | Descriptor: | Spike glycoprotein, antibody H, antibody L | | Authors: | Wang, X, Zhao, J, Wang, Z, Wang, Y, Zeng, J. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | MERS S ectodomain trimer in complex with neutralizing antibody 6516
to be published
|
|
6KVH
 
 | | The mutant crystal structure of endo-polygalacturonase (T284A) from Talaromyces leycettanus JCM 12802 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose, endo-polygalacturonase | | Authors: | Tu, T, Wang, Z, Luo, H, Yao, B. | | Deposit date: | 2019-09-04 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Insights into the Mechanisms Underlying the Kinetic Stability of GH28 Endo-Polygalacturonase.
J.Agric.Food Chem., 69, 2021
|
|
6Y6I
 
 | | lipopolysaccharide outer core galactosyltransferase WaaB and UDP complex | | Descriptor: | Lipopolysaccharide 1,6-galactosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhang, Z.Y, Ashworth, G, Wang, Z.S, Zhu, X.F, Dong, C.J. | | Deposit date: | 2020-02-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural insights into the lipopolysaccharide outer core galactosyltransferase WaaB
To Be Published
|
|
6Y6G
 
 | | lipopolysaccharide outer core galactosyltransferase WaaB apo form | | Descriptor: | Lipopolysaccharide 1,6-galactosyltransferase | | Authors: | Zhang, Z.Y, Ashworth, G, Wang, Z.S, Zhu, X.F, Dong, C.J. | | Deposit date: | 2020-02-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural insights into the lipopolysaccharide outer core galactosyltransferase WaaB
To Be Published
|
|
4DUM
 
 | | Co-crystal structure of eIF4E with inhibitor | | Descriptor: | (4-{7-[2-(4-chlorophenoxy)ethyl]-2-(methylamino)-6-oxo-6,7-dihydro-1H-purin-8-yl}phenyl)phosphonic acid, 1,2-ETHANEDIOL, Eukaryotic translation initiation factor 4E | | Authors: | Min, X, Johnstone, S, Walker, N, Wang, Z. | | Deposit date: | 2012-02-22 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure-Guided Design, Synthesis, and Evaluation of Guanine-Derived Inhibitors of the eIF4E mRNA-Cap Interaction.
J.Med.Chem., 55, 2012
|
|
3NK7
 
 | | Structure of the Nosiheptide-resistance methyltransferase S-adenosyl-L-methionine Complex | | Descriptor: | 23S rRNA methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Yang, H, Wang, Z, Shen, Y, Wang, P, Murchie, A, Xu, Y. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Nosiheptide-Resistance Methyltransferase of Streptomyces actuosus
Biochemistry, 49, 2010
|
|
