9INP
 
 | | Human Pin1 (Peptidyl-prolyl cis-trans isomerase) catalytic domain in complex with a covalent inhibitor | | Descriptor: | CITRIC ACID, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ~{N}-[(3~{R})-1,1-bis(oxidanylidene)thiolan-3-yl]-2-chloranyl-~{N}-(2,2-dimethylpropyl)-5-nitro-pyrimidin-4-amine | | Authors: | Wang, X.Y, Tian, M.Z, Zhou, J, Xu, B.L. | | Deposit date: | 2024-07-08 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Discovery of Novel Pyrimidine Derivatives as Human Pin1 Covalent Inhibitors.
Acs Med.Chem.Lett., 16, 2025
|
|
8HLR
 
 | | Human ADP-ribosyltransferase 1 (PARP1) catalytic domain bound to Fluzoparib (SHR3162) | | Descriptor: | Fluzoparib, Poly [ADP-ribose] polymerase 1, processed C-terminus, ... | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2022-11-30 | | Release date: | 2024-05-01 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Engaging an engineered PARP-2 catalytic domain mutant to solve the complex structures harboring approved drugs for structure analyses.
Bioorg.Chem., 160, 2025
|
|
8HLQ
 
 | |
9J1H
 
 | | The binary complex structure of F2Y224-FtmOx1 mutant with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, COBALT (II) ION, GLYCEROL, ... | | Authors: | Wang, X.Y, Wang, J, Yan, W.P. | | Deposit date: | 2024-08-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterizing Y224 conformational flexibility in FtmOx1-catalysis using 19 F NMR spectroscopy.
Catalysis Science And Technology, 15, 2025
|
|
9J1I
 
 | |
8JNY
 
 | | Mutated human ADP-ribosyltransferase 2 (PARP2) catalytic domain bound to a pyrazolopyrimidine carboxamide inhibitor | | Descriptor: | 6-methylpyrazolo[1,5-a]pyrimidine-3-carboxamide, Poly [ADP-ribose] polymerase 2 | | Authors: | Wang, X.Y, Wang, C.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Employing a Highly Potent Fluorescence Probe to Discover a PARP-1/2 Binder and the Complex Structures Analysis.
Chemmedchem, 2025
|
|
8JNZ
 
 | | Human ADP-ribosyltransferase 1 (PARP1) catalytic domain bound to a pyrazolopyrimidine carboxamide inhibitor | | Descriptor: | 6-methylpyrazolo[1,5-a]pyrimidine-3-carboxamide, Poly [ADP-ribose] polymerase 1, processed C-terminus, ... | | Authors: | Wang, X.Y, Wang, C.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Employing a Highly Potent Fluorescence Probe to Discover a PARP-1/2 Binder and the Complex Structures Analysis.
Chemmedchem, 2025
|
|
6LW2
 
 | | The N-arylsulfonyl-indole-2-carboxamide-based inhibitors against fructose-1,6-bisphosphatase | | Descriptor: | 7-chloranyl-4-[(3-methoxyphenyl)amino]-N-(4-methoxyphenyl)sulfonyl-1-methyl-indole-2-carboxamide, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2020-02-07 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of N -Arylsulfonyl-Indole-2-Carboxamide Derivatives as Potent, Selective, and Orally Bioavailable Fructose-1,6-Bisphosphatase Inhibitors-Design, Synthesis, In Vivo Glucose Lowering Effects, and X-ray Crystal Complex Analysis.
J.Med.Chem., 63, 2020
|
|
8W90
 
 | | crystal structure of CD4-D1D2 with Nb457 | | Descriptor: | NB457, T-cell surface glycoprotein CD4 | | Authors: | Wang, X.Y, Wang, Y.X. | | Deposit date: | 2023-09-04 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure of CD4-D1D2 domain and nanobody Nb457 at 1.78 Angstroms resolution
To Be Published
|
|
8HE8
 
 | | Human ADP-ribosyltransferase 2 (PARP2) catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[4-fluoranyl-3-(3-oxidanylidene-4-pentan-3-yl-piperazin-1-yl)carbonyl-phenyl]methyl]quinazoline-2,4-dione, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Wang, X.Y, Xu, B.L, Zhou, J. | | Deposit date: | 2022-11-07 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Discovery of Quinazoline-2,4(1 H ,3 H )-dione Derivatives Containing a Piperizinone Moiety as Potent PARP-1/2 Inhibitors─Design, Synthesis, In Vivo Antitumor Activity, and X-ray Crystal Structure Analysis.
J.Med.Chem., 66, 2023
|
|
8HE7
 
 | | ADP-ribosyltransferase 1 (PARP1) catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[4-fluoranyl-3-(3-oxidanylidene-4-pentan-3-yl-piperazin-1-yl)carbonyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1, processed C-terminus, ... | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2022-11-07 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Quinazoline-2,4(1 H ,3 H )-dione Derivatives Containing a Piperizinone Moiety as Potent PARP-1/2 Inhibitors─Design, Synthesis, In Vivo Antitumor Activity, and X-ray Crystal Structure Analysis.
J.Med.Chem., 66, 2023
|
|
8HKN
 
 | |
8HLJ
 
 | |
8HKO
 
 | |
8HKS
 
 | | Mutated human ADP-ribosyltransferase 2 (PARP2) catalytic domain bound to Pamiparib(BGB-290) | | Descriptor: | (2R)-14-fluoro-2-methyl-6,9,10,19-tetrazapentacyclo[14.2.1.02,6.08,18.012,17]nonadeca-1(18),8,12(17),13,15-pentaen-11-one, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2022-11-28 | | Release date: | 2024-05-01 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Engaging an engineered PARP-2 catalytic domain mutant to solve the complex structures harboring approved drugs for structure analyses.
Bioorg.Chem., 160, 2025
|
|
7EZP
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3-(3-hydroxy-3-oxopropyl)-5-(2-methylpropyl)-7-nitro-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7EZF
 
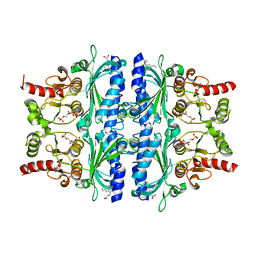 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 7-chloranyl-5-ethyl-3-(3-hydroxy-3-oxopropyl)-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7EZR
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 5-ethyl-7-nitro-3-[3-oxidanylidene-3-(thiophen-2-ylsulfonylamino)propyl]-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
3BGG
 
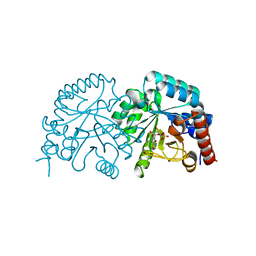 | | Crystal structure of Human Orotidine 5'-monophosphate Decarboxylase complexed with BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Uridine 5'-monophosphate synthase | | Authors: | Liu, Y, Tang, H.L, Wang, X.Y, Kotra, L.P, Pai, E.F. | | Deposit date: | 2007-11-26 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of Human Orotidine 5'-monophosphate Decarboxylase complexed with BMP
To be Published
|
|
8WUL
 
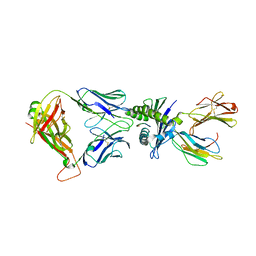 | | Crystal structure of affinity enhanced TCR in complex with HLA-A*11:01 bound to KRAS-G12V peptide (VVGAVGVGK) | | Descriptor: | Beta-2-microglobulin, KRAS-G12V nonamer peptide, MHC class I antigen, ... | | Authors: | Zhang, M.Y, Luo, L.J, Xu, W, Guan, F.H, Wang, X.Y, Zhu, P, Zhang, J.H, Zhou, X.Y, Wang, F, Ye, S. | | Deposit date: | 2023-10-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Identification and affinity enhancement of T-cell receptor targeting a KRAS G12V cancer neoantigen.
Commun Biol, 7, 2024
|
|
8WTE
 
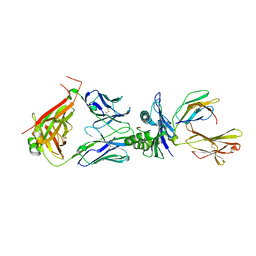 | | Crystal structure of TCR in complex with HLA-A*11:01 bound to KRAS-G12V peptide (VVGAVGVGK) | | Descriptor: | Beta-2-microglobulin, KRAS-G12V nonamer peptide, MHC class I antigen (Fragment), ... | | Authors: | Zhang, M.Y, Luo, L.J, Xu, W, Guan, F.H, Wang, X.Y, Zhu, P, Zhang, J.H, Zhou, X.Y, Wang, F, Ye, S. | | Deposit date: | 2023-10-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Identification and affinity enhancement of T-cell receptor targeting a KRAS G12V cancer neoantigen.
Commun Biol, 7, 2024
|
|
7WSB
 
 | | The ternary complex structure of FtmOx1 with a-ketoglutarate and 13-oxo-fumitremorgin B | | Descriptor: | 13-Oxofumitremorgin B, 2-OXOGLUTARIC ACID, COBALT (II) ION, ... | | Authors: | Wang, J, Wang, X.Y, Wang, Y.Y, Yan, W.P. | | Deposit date: | 2022-01-28 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Dissecting the Mechanism of the Nonheme Iron Endoperoxidase FtmOx1 Using Substrate Analogues.
Jacs Au, 2, 2022
|
|
8KHQ
 
 | | Bifunctional sulfoxide synthase OvoA_Th2 in complex with histidine and cysteine | | Descriptor: | 5-histidylcysteine sulfoxide synthase/putative 4-mercaptohistidine N1-methyltranferase, COBALT (II) ION, CYSTEINE, ... | | Authors: | Wang, J, Ye, K, Wang, X.Y, Yan, W.P. | | Deposit date: | 2023-08-22 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Biochemical and Structural Characterization of OvoA Th2 : A Mononuclear Nonheme Iron Enzyme from Hydrogenimonas thermophila for Ovothiol Biosynthesis.
Acs Catalysis, 13, 2023
|
|
