8K5Q
 
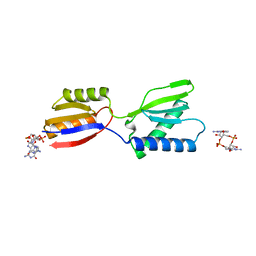 | | Crystal structure of YajQ STM0435 with c-di-GMP | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), YajQ | | 著者 | Dai, Y, Zhang, M, Wang, W, Li, B. | | 登録日 | 2023-07-23 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | A c-di-GMP binding effector STM0435 modulates flagellar motility and pathogenicity in Salmonella
Virulence, 15, 2024
|
|
4B5V
 
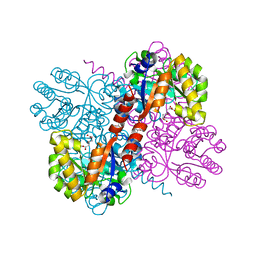 | | Crystal structures of divalent metal dependent pyruvate aldolase, HpaI, in complex with 4-hydroxyl-2-ketoheptane-1,7-dioate | | 分子名称: | (4R)-4-oxidanyl-2-oxidanylidene-heptanedioic acid, 4-HYDROXY-2-OXO-HEPTANE-1,7-DIOATE ALDOLASE, GLYCEROL, ... | | 著者 | Coincon, M, Wang, W, Seah, S.Y.K, Sygusch, J. | | 登録日 | 2012-08-07 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.041 Å) | | 主引用文献 | Crystal Structure of Reaction Intermediates in Pyruvate Class II Aldolase: Substrate Cleavage, Enolate Stabilization and Substrate Specificity
J.Biol.Chem., 287, 2012
|
|
4B5U
 
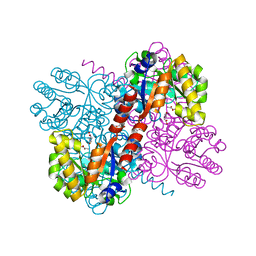 | | Crystal structures of divalent metal dependent pyruvate aldolase, HpaI, in complex with pyruvate and succinic semialdehyde | | 分子名称: | 4-HYDROXY-2-OXO-HEPTANE-1,7-DIOATE ALDOLASE, 4-oxobutanoic acid, COBALT (II) ION, ... | | 著者 | Coincon, M, Wang, W, Seah, S.Y.K, Sygusch, J. | | 登録日 | 2012-08-07 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.913 Å) | | 主引用文献 | Crystal Structure of Reaction Intermediates in Pyruvate Class II Aldolase: Substrate Cleavage, Enolate Stabilization and Substrate Specificity
J.Biol.Chem., 287, 2012
|
|
4B5T
 
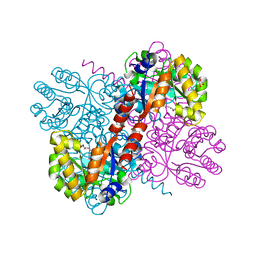 | | Crystal structures of divalent metal dependent pyruvate aldolase, HpaI, in complex with ketobutyrate | | 分子名称: | 2-KETOBUTYRIC ACID, 4-HYDROXY-2-OXO-HEPTANE-1,7-DIOATE ALDOLASE, COBALT (II) ION, ... | | 著者 | Coincon, M, Wang, W, Seah, S.Y.K, Sygusch, J. | | 登録日 | 2012-08-07 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.923 Å) | | 主引用文献 | Crystal Structure of Reaction Intermediates in Pyruvate Class II Aldolase: Substrate Cleavage, Enolate Stabilization and Substrate Specificity
J.Biol.Chem., 287, 2012
|
|
4B5W
 
 | | Crystal structures of divalent metal dependent pyruvate aldolase R70A mutant, HpaI, in complex with pyruvate | | 分子名称: | 4-HYDROXY-2-OXO-HEPTANE-1,7-DIOATE ALDOLASE, CALCIUM ION, COBALT (II) ION, ... | | 著者 | Coincon, M, Wang, W, Seah, S.Y.K, Sygusch, J. | | 登録日 | 2012-08-07 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.792 Å) | | 主引用文献 | Crystal Structure of Reaction Intermediates in Pyruvate Class II Aldolase: Substrate Cleavage, Enolate Stabilization and Substrate Specificity
J.Biol.Chem., 287, 2012
|
|
8I79
 
 | |
4B5S
 
 | | Crystal structures of divalent metal dependent pyruvate aldolase, HpaI, in complex with pyruvate | | 分子名称: | 4-HYDROXY-2-OXO-HEPTANE-1,7-DIOATE ALDOLASE, COBALT (II) ION, D-Glyceraldehyde, ... | | 著者 | Coincon, M, Wang, W, Seah, S.Y.K, Sygusch, J. | | 登録日 | 2012-08-07 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Crystal Structure of Reaction Intermediates in Pyruvate Class II Aldolase: Substrate Cleavage, Enolate Stabilization and Substrate Specificity
J.Biol.Chem., 287, 2012
|
|
7KED
 
 | | RNA polymerase II elongation complex with unnatural base dTPT3 | | 分子名称: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | 著者 | Oh, J, Wang, W, Wang, D. | | 登録日 | 2020-10-10 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Transcriptional processing of an unnatural base pair by eukaryotic RNA polymerase II.
Nat.Chem.Biol., 17, 2021
|
|
4DSO
 
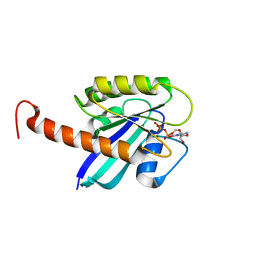 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | 分子名称: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, BENZAMIDINE, GLYCEROL, ... | | 著者 | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | 登録日 | 2012-02-19 | | 公開日 | 2012-04-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DSU
 
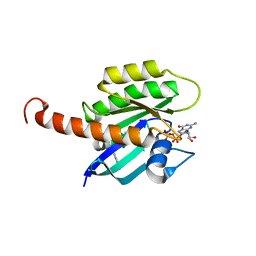 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | 分子名称: | BENZIMIDAZOLE, GTPase KRas, isoform 2B, ... | | 著者 | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | 登録日 | 2012-02-19 | | 公開日 | 2012-04-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DSN
 
 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | 分子名称: | 1,2-ETHANEDIOL, GTPase KRas, isoform 2B, ... | | 著者 | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | 登録日 | 2012-02-19 | | 公開日 | 2012-04-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3LF5
 
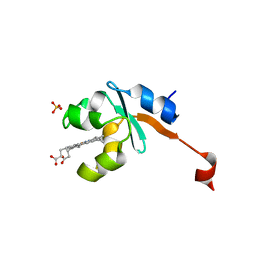 | | Structure of Human NADH cytochrome b5 oxidoreductase (Ncb5or) b5 Domain to 1.25A Resolution | | 分子名称: | Cytochrome b5 reductase 4, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | 著者 | Deng, B, Parthasarathy, S, Wang, W, Gibney, B.R, Battaile, K.P, Lovell, S, Benson, D.R, Zhu, H. | | 登録日 | 2010-01-15 | | 公開日 | 2010-07-14 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Study of the individual cytochrome b5 and cytochrome b5 reductase domains of Ncb5or reveals a unique heme pocket and a possible role of the CS domain.
J.Biol.Chem., 285, 2010
|
|
3L82
 
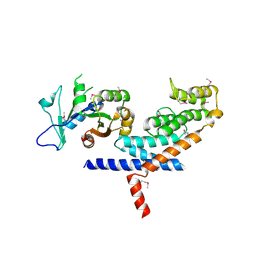 | | X-ray Crystal structure of TRF1 and Fbx4 complex | | 分子名称: | F-box only protein 4, Telomeric repeat-binding factor 1 | | 著者 | Zeng, Z.X, Wang, W, Yang, Y.T, Chen, Y, Yang, X.M, Diehl, J.A, Liu, X.D, Lei, M. | | 登録日 | 2009-12-29 | | 公開日 | 2010-03-09 | | 最終更新日 | 2013-09-25 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Basis of Selective Ubiquitination of TRF1 by SCF(Fbx4)
Dev.Cell, 18, 2010
|
|
8GQR
 
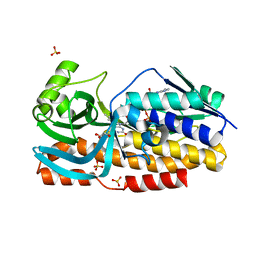 | | Crystal structure of VioD with FAD | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | 著者 | Ran, T, Wang, W, Xu, M. | | 登録日 | 2022-08-30 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for substrate binding and catalytic mechanism of the key enzyme VioD in the violacein synthesis pathway.
Proteins, 91, 2023
|
|
8H0M
 
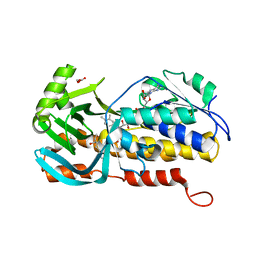 | | Crystal structure of VioD | | 分子名称: | (2S)-2-ethylhexan-1-ol, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | 著者 | Xu, M, Ran, T, Wang, W. | | 登録日 | 2022-09-29 | | 公開日 | 2023-04-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.702 Å) | | 主引用文献 | Structural basis for substrate binding and catalytic mechanism of the key enzyme VioD in the violacein synthesis pathway.
Proteins, 91, 2023
|
|
8GOY
 
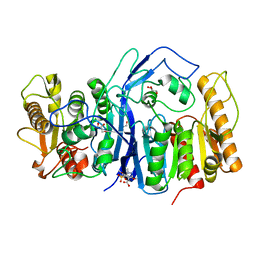 | | SulE P44R | | 分子名称: | 5-[(4,6-dimethoxypyrimidin-2-yl)carbamoylsulfamoyl]-1-methyl-pyrazole-4-carboxylic acid, Alpha/beta fold hydrolase, GLYCEROL | | 著者 | Liu, B, He, J, Ran, T, Wang, W. | | 登録日 | 2022-08-25 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.784 Å) | | 主引用文献 | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8GP0
 
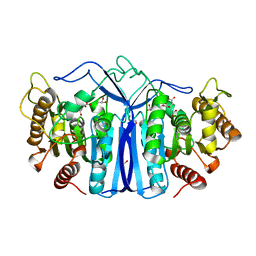 | | crystal structure of SulE | | 分子名称: | Alpha/beta fold hydrolase, CITRIC ACID, GLYCEROL | | 著者 | Liu, B, Ran, T, wang, W, He, J. | | 登録日 | 2022-08-25 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8GOL
 
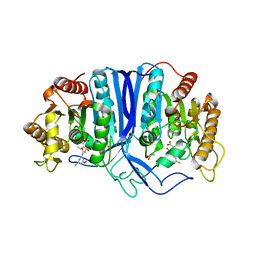 | | crystal structure of SulE | | 分子名称: | 2-[(4-chloranyl-6-methoxy-pyrimidin-2-yl)carbamoylsulfamoyl]benzoic acid, Alpha/beta fold hydrolase, GLYCEROL | | 著者 | Liu, B, Ran, T, Wang, W, He, J. | | 登録日 | 2022-08-25 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
3MXN
 
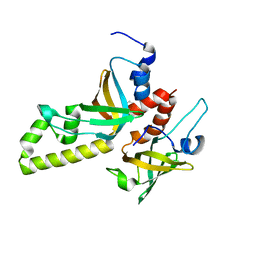 | | Crystal structure of the RMI core complex | | 分子名称: | BENZAMIDINE, RecQ-mediated genome instability protein 1, RecQ-mediated genome instability protein 2 | | 著者 | Hoadley, K.A, Xu, D, Xue, Y, Satyshur, K.A, Wang, W, Keck, J.L. | | 登録日 | 2010-05-07 | | 公開日 | 2010-09-15 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structure and cellular roles of the RMI core complex from the bloom syndrome dissolvasome.
Structure, 18, 2010
|
|
8HEH
 
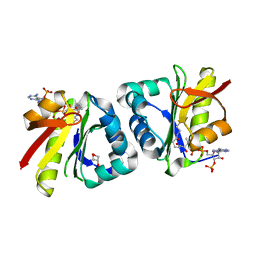 | | Crystal structure of GCN5-related N-acetyltransferase 05790 | | 分子名称: | COENZYME A, GLYCEROL, GNAT family N-acetyltransferase | | 著者 | Xu, M.X, Ran, T.T, Wang, W. | | 登録日 | 2022-11-08 | | 公開日 | 2022-12-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of prodigiosin binding protein PgbP, a GNAT family protein, in Serratia marcescens FS14.
Biochem.Biophys.Res.Commun., 640, 2022
|
|
3NOJ
 
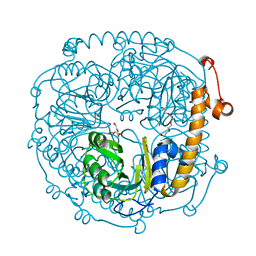 | | The structure of HMG/CHA aldolase from the protocatechuate degradation pathway of Pseudomonas putida | | 分子名称: | 4-carboxy-4-hydroxy-2-oxoadipate aldolase/oxaloacetate decarboxylase, MAGNESIUM ION, PYRUVIC ACID, ... | | 著者 | Kimber, M.S, Wang, W, Mazurkewich, S, Seah, S.Y.K. | | 登録日 | 2010-06-25 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Structural and Kinetic Characterization of 4-Hydroxy-4-methyl-2-oxoglutarate/4-Carboxy-4-hydroxy-2-oxoadipate Aldolase, a Protocatechuate Degradation Enzyme Evolutionarily Convergent with the HpaI and DmpG Pyruvate Aldolases.
J.Biol.Chem., 285, 2010
|
|
3MWP
 
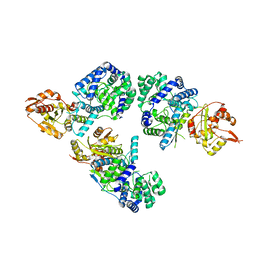 | | Nucleoprotein structure of lassa fever virus | | 分子名称: | Nucleoprotein, ZINC ION | | 著者 | Qi, X, Lan, S, Wang, W, Schelde, L.M, Dong, H, Wallat, G, Liang, Y, Ly, H, Dong, C, Scottish Structural Proteomics Facility (SSPF) | | 登録日 | 2010-05-06 | | 公開日 | 2010-12-01 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.795 Å) | | 主引用文献 | Cap binding and immune evasion revealed by Lassa nucleoprotein structure.
Nature, 468, 2010
|
|
3MX5
 
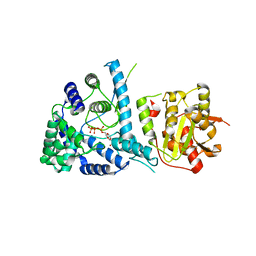 | | Lassa fever virus nucleoprotein complexed with UTP | | 分子名称: | Nucleoprotein, URIDINE 5'-TRIPHOSPHATE, ZINC ION | | 著者 | Qi, X, Lan, S, Wang, W, Schelde, L.M, Dong, H, Wallat, G, Liang, Y, Ly, H, Dong, C, Scottish Structural Proteomics Facility (SSPF) | | 登録日 | 2010-05-06 | | 公開日 | 2010-12-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.903 Å) | | 主引用文献 | Cap binding and immune evasion revealed by Lassa nucleoprotein structure.
Nature, 468, 2010
|
|
3MWT
 
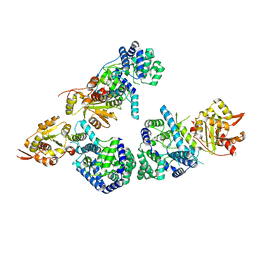 | | Crystal structure of Lassa fever virus nucleoprotein in complex with Mn2+ | | 分子名称: | MANGANESE (II) ION, Nucleoprotein, ZINC ION | | 著者 | Qi, X, Lan, S, Wang, W, Schelde, L.M, Dong, H, Wallat, G, Liang, Y, Ly, H, Dong, C, Scottish Structural Proteomics Facility (SSPF) | | 登録日 | 2010-05-06 | | 公開日 | 2010-12-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.982 Å) | | 主引用文献 | Cap binding and immune evasion revealed by Lassa nucleoprotein structure.
Nature, 468, 2010
|
|
3MX2
 
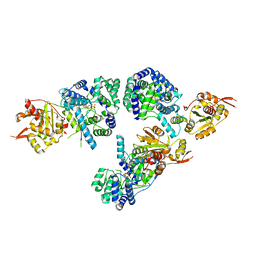 | | Lassa fever virus Nucleoprotein complexed with dTTP | | 分子名称: | Nucleoprotein, THYMIDINE-5'-TRIPHOSPHATE, ZINC ION | | 著者 | Qi, X, Lan, S, Wang, W, Schelde, L.M, Dong, H, Wallat, G, Liang, Y, Ly, H, Dong, C, Scottish Structural Proteomics Facility (SSPF) | | 登録日 | 2010-05-06 | | 公開日 | 2010-12-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.983 Å) | | 主引用文献 | Cap binding and immune evasion revealed by Lassa nucleoprotein structure.
Nature, 468, 2010
|
|
