7YIL
 
 | |
7YOQ
 
 | | Recj2 and CMP complex from Methanocaldococcus jannaschii | | Descriptor: | [5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)furan-2-yl]methyl dihydrogen phosphate, nuclease RecJ2 | | Authors: | Wang, W.W, Liu, X.P. | | Deposit date: | 2022-08-02 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.265 Å) | | Cite: | The structure of nuclease RecJ2 from Methanocaldococcus jannaschii
To Be Published
|
|
7YOS
 
 | |
6IVE
 
 | |
2VV5
 
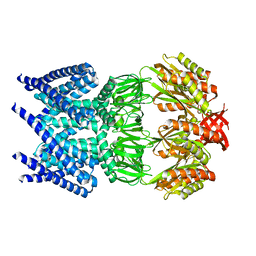 | | The open structure of MscS | | Descriptor: | SMALL-CONDUCTANCE MECHANOSENSITIVE CHANNEL | | Authors: | Wang, W, Dong, C, Johnson, K.A, Naismith, J.H. | | Deposit date: | 2008-06-03 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The Structure of an Open Form of an E. Coli Mechanosensitive Channel at 3.45 A Resolution.
Science, 321, 2008
|
|
1S4M
 
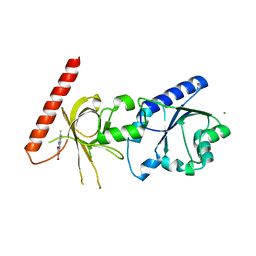 | | Crystal structure of flavin binding to FAD synthetase from Thermotoga maritina | | Descriptor: | LUMICHROME, MAGNESIUM ION, riboflavin kinase/FMN adenylyltransferase | | Authors: | Wang, W, Kim, R, Yokota, H, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-16 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of flavin binding to FAD synthetase of Thermotoga maritima
Proteins, 58, 2005
|
|
1L7P
 
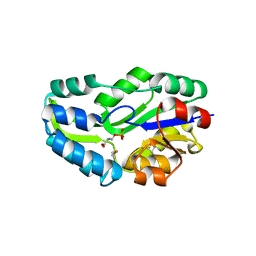 | | SUBSTRATE BOUND PHOSPHOSERINE PHOSPHATASE COMPLEX STRUCTURE | | Descriptor: | PHOSPHATE ION, PHOSPHOSERINE, PHOSPHOSERINE PHOSPHATASE | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1L7N
 
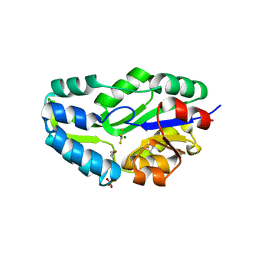 | | TRANSITION STATE ANALOGUE OF PHOSPHOSERINE PHOSPHATASE (ALUMINUM FLUORIDE COMPLEX) | | Descriptor: | ALUMINUM FLUORIDE, MAGNESIUM ION, PHOSPHOSERINE PHOSPHATASE, ... | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1L7O
 
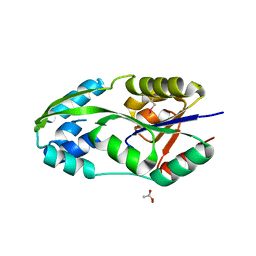 | | CRYSTAL STRUCTURE OF PHOSPHOSERINE PHOSPHATASE IN APO FORM | | Descriptor: | ACETIC ACID, PHOSPHOSERINE PHOSPHATASE, ZINC ION | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
4EB3
 
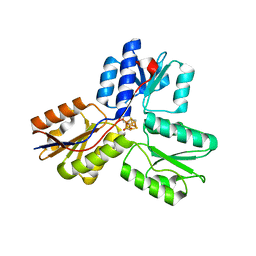 | | Crystal structure of IspH in complex with iso-HMBPP | | Descriptor: | 3-(hydroxymethyl)but-3-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Wang, W, Wang, K, Span, I, Bacher, A, Groll, M, Oldfield, E. | | Deposit date: | 2012-03-23 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Are free radicals involved in IspH catalysis? An EPR and crystallographic investigation.
J.Am.Chem.Soc., 134, 2012
|
|
4Z8I
 
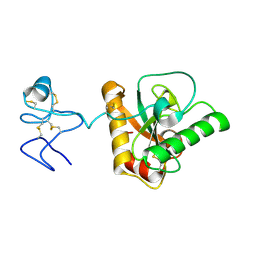 | | Crystal structure of Branchiostoma belcheri tsingtauense peptidoglycan recognition protein 3 | | Descriptor: | ZINC ION, peptidoglycan recognition protein 3 | | Authors: | Wang, W.J, Cheng, W, Jiang, Y.L, Luo, M, Cao, D.D, Chi, C.B, Yang, H.B, Chen, Y, Zhou, C.Z. | | Deposit date: | 2015-04-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Activity Augmentation of Amphioxus Peptidoglycan Recognition Protein BbtPGRP3 via Fusion with a Chitin Binding Domain
Plos One, 10, 2015
|
|
4ZXM
 
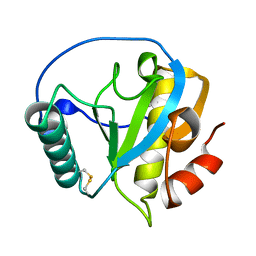 | | Crystal structure of PGRP domain from Branchiostoma belcheri tsingtauense peptidoglycan recognition protein 3 | | Descriptor: | PGRP domain of peptidoglycan recognition protein 3 | | Authors: | Wang, W.J, Cheng, W, Jiang, Y.L, Yu, H.M, Luo, M. | | Deposit date: | 2015-05-20 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Activity Augmentation of Amphioxus Peptidoglycan Recognition Protein BbtPGRP3 via Fusion with a Chitin Binding Domain
Plos One, 10, 2015
|
|
6NTV
 
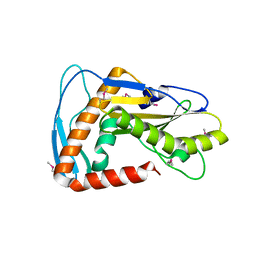 | | SFTSV L endonuclease domain | | Descriptor: | RNA polymerase | | Authors: | Wang, W, Amarasinghe, G.K. | | Deposit date: | 2019-01-30 | | Release date: | 2020-01-08 | | Last modified: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Cap-Snatching SFTSV Endonuclease Domain Is an Antiviral Target.
Cell Rep, 30, 2020
|
|
5VA3
 
 | |
5VA2
 
 | |
5ZND
 
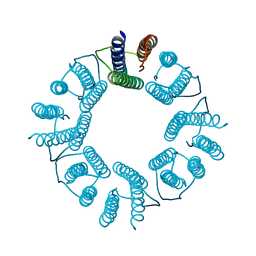 | | 8-mer nanotube derived from 24-mer rHuHF nanocage | | Descriptor: | Ferritin heavy chain | | Authors: | Wang, W.M, Wang, L.L, Zang, J.C, Chen, H, Zhao, G.H, Wang, H.F. | | Deposit date: | 2018-04-09 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Selective Elimination of the Key Subunit Interfaces Facilitates Conversion of Native 24-mer Protein Nanocage into 8-mer Nanorings.
J. Am. Chem. Soc., 140, 2018
|
|
5VA1
 
 | |
1PWJ
 
 | | Structure of the Monomeric 8-kDa Dynein Light Chain and Mechanism of Domain Swapped Dimer Assembly | | Descriptor: | dynein light chain-2 | | Authors: | Wang, W, Lo, K.W.-H, Kan, H.-M, Fan, J.-S, Zhang, M. | | Deposit date: | 2003-07-02 | | Release date: | 2003-10-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Monomeric 8-kDa Dynein Light Chain and Mechanism of the Domain-swapped Dimer Assembly
J.Biol.Chem., 278, 2003
|
|
5GJT
 
 | | Crystal structure of H1 hemagglutinin from A/Washington/05/2011 in complex with a neutralizing antibody 3E1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, heavy chain of human neutralizing antibody 3E1, ... | | Authors: | Wang, W, Zhang, T, Ding, J. | | Deposit date: | 2016-07-01 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Human antibody 3E1 targets the HA stem region of H1N1 and H5N6 influenza A viruses
Nat Commun, 7, 2016
|
|
5GJS
 
 | | Crystal structure of H1 hemagglutinin from A/California/04/2009 in complex with a neutralizing antibody 3E1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Wang, W, Zhang, T, Ding, J. | | Deposit date: | 2016-07-01 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Human antibody 3E1 targets the HA stem region of H1N1 and H5N6 influenza A viruses
Nat Commun, 7, 2016
|
|
1PWK
 
 | | Structure of the Monomeric 8-kDa Dynein Light Chain and Mechanism of Domain Swapped Dimer Assembly | | Descriptor: | dynein light chain-2 | | Authors: | Wang, W, Lo, K.W.-H, Kan, H.-M, Fan, J.-S, Zhang, M. | | Deposit date: | 2003-07-02 | | Release date: | 2003-10-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Monomeric 8-kDa Dynein Light Chain and Mechanism of the Domain-swapped Dimer Assembly
J.Biol.Chem., 278, 2003
|
|
1OYX
 
 | | CRYSTAL STRUCTURE OF 3-MBT REPEATS OF LETHAL (3) MALIGNANT BRAIN TUMOR (SELENO-MET) AT 1.85 ANGSTROM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lethal(3)malignant brain tumor-like protein, SULFATE ION | | Authors: | Wang, W.K, Tereshko, V, Boccuni, P, MacGrogan, D, Nimer, S.D, Patel, D.J. | | Deposit date: | 2003-04-07 | | Release date: | 2003-08-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Malignant brain tumor repeats: a three-leaved propeller architecture with ligand/peptide binding pockets.
Structure, 11, 2003
|
|
7VP8
 
 | | Crystal structure of ferritin from Ureaplasma urealyticum | | Descriptor: | CHLORIDE ION, FE (III) ION, Ferritin-like diiron domain-containing protein | | Authors: | Wang, W, Liu, X, Wang, Y, Fu, D, Wang, H. | | Deposit date: | 2021-10-15 | | Release date: | 2022-08-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Distinct structural characteristics define a new subfamily of Mycoplasma ferritin
Chin.Chem.Lett., 33, 2022
|
|
3VPV
 
 | |
6KE4
 
 | | ABloop reengineered Ferritin Nanocage | | Descriptor: | CALCIUM ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Wang, W.M, Wang, H.F. | | Deposit date: | 2019-07-03 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AB loop engineered ferritin nanocages for drug loading under benign experimental conditions.
Chem.Commun.(Camb.), 55, 2019
|
|
