8H45
 
 | | Blasnase-T13A/P55N | | Descriptor: | FORMIC ACID, L-asparaginase, MAGNESIUM ION | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H48
 
 | | Blasnase-T13A/P55F with L-asn | | Descriptor: | ASPARAGINE, FORMIC ACID, L-asparaginase, ... | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H49
 
 | | Blasnase-M57P | | Descriptor: | FORMIC ACID, L-asparaginase, MAGNESIUM ION | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H4A
 
 | | Blasnase-T13A/M57P | | Descriptor: | FORMIC ACID, L-asparaginase, MAGNESIUM ION | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H4C
 
 | | Blasnase-T13A/M57P | | Descriptor: | FORMIC ACID, L-asparaginase, MAGNESIUM ION | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H4G
 
 | | Blasnase-T13A/M57N | | Descriptor: | FORMIC ACID, L-asparaginase, MAGNESIUM ION | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H4F
 
 | | Blasnase-T13A/P55F with D-asn | | Descriptor: | D-ASPARAGINE, FORMIC ACID, L-asparaginase, ... | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8I79
 
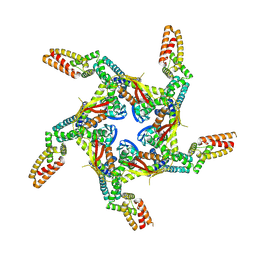 | | Cryo-EM structure of KCTD7 in complex with Cullin3 | | Descriptor: | BTB/POZ domain-containing protein KCTD7, Cullin-3 | | Authors: | Jiang, W, Wang, W, Zheng, S. | | Deposit date: | 2023-01-31 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for the ubiquitination of G protein beta gamma subunits by KCTD5/Cullin3 E3 ligase.
Sci Adv, 9, 2023
|
|
8GQR
 
 | | Crystal structure of VioD with FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Ran, T, Wang, W, Xu, M. | | Deposit date: | 2022-08-30 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for substrate binding and catalytic mechanism of the key enzyme VioD in the violacein synthesis pathway.
Proteins, 91, 2023
|
|
7L31
 
 | | Cyro-EM structure of human Glycine Receptor alpha2-beta heteromer, strychnine bound state, 3.8 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alpha-2, Glycine receptor subunit beta,Green fluorescent protein, ... | | Authors: | Yu, H, Wang, W. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Characterization of the subunit composition and structure of adult human glycine receptors
Neuron, 109, 2021
|
|
7KUY
 
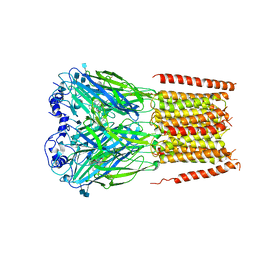 | |
8JKB
 
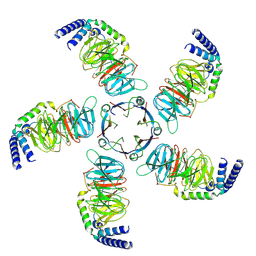 | | Cryo-EM structure of KCTD5 in complex with Gbeta gamma subunits | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | | Authors: | Zheng, S, Jiang, W, Wang, W, Kong, Y. | | Deposit date: | 2023-06-01 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural basis for the ubiquitination of G protein beta gamma subunits by KCTD5/Cullin3 E3 ligase.
Sci Adv, 9, 2023
|
|
8IVM
 
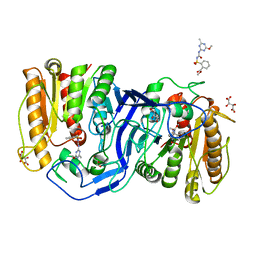 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-28 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IVS
 
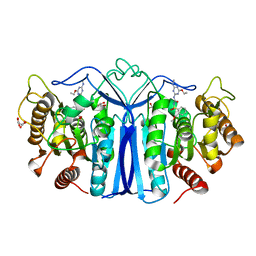 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-28 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8J7J
 
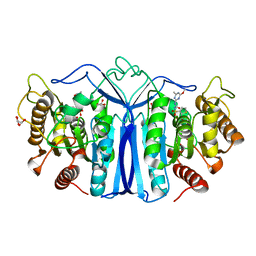 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-04-27 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IW3
 
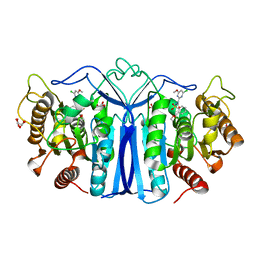 | | crystal structure of SulE mutant | | Descriptor: | 2-[[[[(4-CHLORO-6-METHOXY-2-PYRIMIDINYL)AMINO]CARBONYL]AMINO]SULFONYL]BENZOIC ACID ETHYL ESTER, Alpha/beta fold hydrolase, GLYCEROL, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8J7K
 
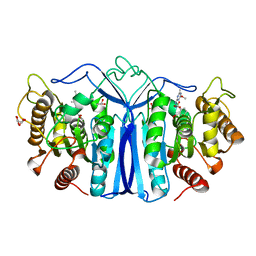 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-04-27 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IW6
 
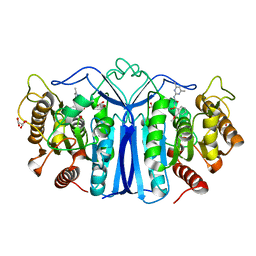 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IVE
 
 | | crystal structure of SulE mutant | | Descriptor: | 2-[[[[(4-CHLORO-6-METHOXY-2-PYRIMIDINYL)AMINO]CARBONYL]AMINO]SULFONYL]BENZOIC ACID ETHYL ESTER, Alpha/beta fold hydrolase, GLYCEROL | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-27 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IVN
 
 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-28 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IVT
 
 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-28 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8J7G
 
 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-04-27 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
6J36
 
 | | crystal structure of Mycoplasma hyopneumoniae Enolase | | Descriptor: | Enolase, GLYCEROL, SULFATE ION | | Authors: | Chen, R, Zhang, S, Gan, R, Xie, X, Feng, Z, Wang, W, Ran, T, Zhang, W, Xiang, Q, Shao, G. | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Featured Species-Specific Loops Are Found in the Crystal Structure ofMhpEno, a Cell Surface Adhesin FromMycoplasma hyopneumoniae.
Front Cell Infect Microbiol, 9, 2019
|
|
1ES1
 
 | | CRYSTAL STRUCTURE OF VAL61HIS MUTANT OF TRYPSIN-SOLUBILIZED FRAGMENT OF CYTOCHROME B5 | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, J, Gan, J.-H, Xia, Z.-X, Wang, Y.-H, Wang, W.-H, Xue, L.-L, Xie, Y, Huang, Z.-X. | | Deposit date: | 2000-04-07 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of recombinant trypsin-solubilized fragment of cytochrome b(5) and the structural comparison with Val61His mutant.
Proteins, 40, 2000
|
|
5Y51
 
 | | Crystal structure of PytH_H230A | | Descriptor: | Pyrethroid hydrolase, SULFATE ION | | Authors: | Xu, D.Q, Ran, T.T, He, J, Wang, W.W. | | Deposit date: | 2017-08-06 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Catalytic Mechanism of a Novel Pyrethroid Hydrolase from Sphingobium faniae JZ-2
To Be Published
|
|
