8FCX
 
 | |
2FEF
 
 | | The Crystal Structure of Protein PA2201 from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, hypothetical protein PA2201 | | Authors: | Cymborowski, M.T, Chruszcz, M, Wang, S, Shumilin, I, Kudritska, M, Evdokimova, E, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-15 | | Release date: | 2006-01-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Protein PA2201 from Pseudomonas aeruginosa
To be Published
|
|
6J66
 
 | | Chondroitin sulfate/dermatan sulfate endolytic 4-O-sulfatase | | Descriptor: | CALCIUM ION, Chondroitin sulfate/dermatan sulfate 4-O-endosulfatase protein | | Authors: | Gu, L, Li, F, Su, T, Wang, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Comparative Study of Two Chondroitin Sulfate/Dermatan Sulfate 4-O-Sulfatases With High Identity.
Front Microbiol, 10, 2019
|
|
2FFS
 
 | | Structure of PR10-allergen-like protein PA1206 from Pseudomonas aeruginosa PAO1 | | Descriptor: | hypothetical protein PA1206 | | Authors: | Zimmerman, M.D, Chruszcz, M, Cymborowski, M.T, Wang, S, Kirillova, O, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-20 | | Release date: | 2006-01-10 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of PR10-Allergen-Like Protein PA1206 From Pseudomonas aeruginosa PAO1
To be Published
|
|
2FEX
 
 | | The Crystal Structure of DJ-1 Superfamily Protein Atu0886 from Agrobacterium tumefaciens | | Descriptor: | GLYCEROL, SULFATE ION, conserved hypothetical protein | | Authors: | Cymborowski, M.T, Wang, S, Chruszcz, M, Shumilin, I, Gu, J, Xu, X, Edwards, A.M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-16 | | Release date: | 2006-01-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of DJ-1 Superfamily Protein Atu0886 from Agrobacterium tumefaciens
To be Published
|
|
2G3B
 
 | | Crystal structure of putative TetR-family transcriptional regulator from Rhodococcus sp. | | Descriptor: | GLYCEROL, putative TetR-family transcriptional regulator | | Authors: | Chruszcz, M, Evdokimova, E, Cymborowski, M, Kagan, O, Wang, S, Koclega, K.D, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-17 | | Release date: | 2006-03-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative TetR-family transcriptional regulator from Rhodococcus sp.
To be Published
|
|
3FZV
 
 | | Crystal structure of PA01 protein, putative LysR family transcriptional regulator from Pseudomonas aeruginosa | | Descriptor: | Probable transcriptional regulator, SULFATE ION | | Authors: | Knapik, A.A, Tkaczuk, K.L, Chruszcz, M, Wang, S, Zimmerman, M.D, Cymborowski, M, Skarina, T, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Bujnicki, J.M, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of PA01 protein, putative LysR family transcriptional regulator from Pseudomonas aeruginosa
To be Published
|
|
3H3M
 
 | | Crystal structure of flagellar protein FliT from Bordetella bronchiseptica | | Descriptor: | Flagellar protein FliT, UNKNOWN | | Authors: | Shumilin, I.A, Wang, S, Chruszcz, M, Xu, X, Le, B, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-16 | | Release date: | 2009-04-28 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of flagellar protein FliT from Bordetella bronchiseptica
To be Published
|
|
5DVV
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with a Triaryl-imine analog 4,4'-(phenylcarbonimidoyl)diphenol | | Descriptor: | 4,4'-(phenylcarbonimidoyl)diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Wright, N.J, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-21 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.505 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DKB
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a 3-methylphenylamino-substituted ethyl triaryl-ethylene derivative 4,4'-(2-{3-[(3-methylphenyl)amino]phenyl}but-1-ene-1,1-diyl)diphenol | | Descriptor: | 4,4'-(2-{3-[(3-methylphenyl)amino]phenyl}but-1-ene-1,1-diyl)diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-03 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DXQ
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 4,4'-[(1s,5s)-bicyclo[3.3.1]non-9-ylidenemethanediyl]diphenol | | Descriptor: | 4,4'-[(1s,5s)-bicyclo[3.3.1]non-9-ylidenemethanediyl]diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-23 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5E19
 
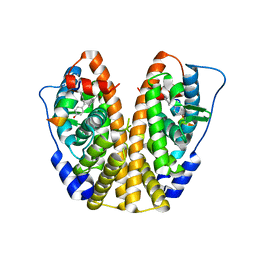 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative methyl {4-[bis(4-hydroxyphenyl)methylidene]cyclohexyl}acetate | | Descriptor: | Estrogen receptor, Nuclear receptor coactivator 2, methyl {4-[bis(4-hydroxyphenyl)methylidene]cyclohexyl}acetate | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-29 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
8WWC
 
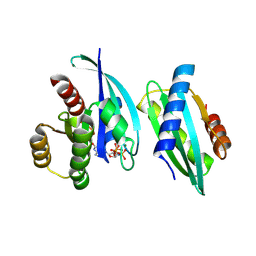 | |
8WX8
 
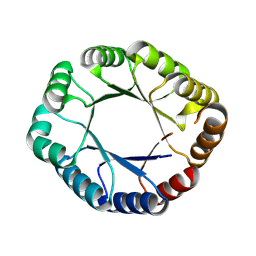 | |
5DKS
 
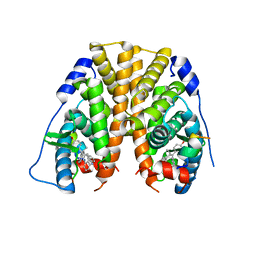 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a 2-naphthylamino-substituted, ethyl, triaryl-ethylene derivative 4,4'-{2-[3-(naphthalen-1-ylamino)phenyl]but-1-ene-1,1-diyl}diphenol | | Descriptor: | 4,4'-{2-[3-(naphthalen-1-ylamino)phenyl]but-1-ene-1,1-diyl}diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-03 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DK9
 
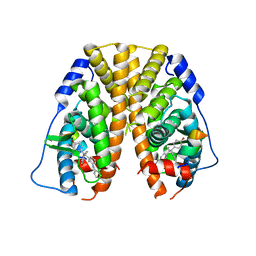 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a phenylamino-substituted ethyl triaryl-ethylene derivative 4,4'-{2-[3-(phenylamino)phenyl]but-1-ene-1,1-diyl}diphenol | | Descriptor: | 4,4'-{2-[3-(phenylamino)phenyl]but-1-ene-1,1-diyl}diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-03 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DLR
 
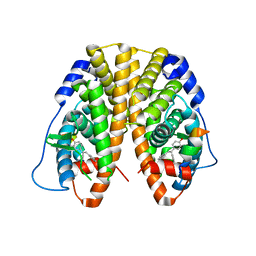 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a triaryl-ethylene compound 4,4'-(2-phenylethene-1,1-diyl)diphenol | | Descriptor: | 4,4'-(2-phenylethene-1,1-diyl)diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-07 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DRJ
 
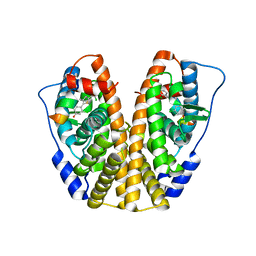 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a dichloro-substituted, 3-methyl 2,5-diarylthiophene-core ligand 4,4'-(3-methylthiene-2,5-diyl)bis(3-chlorophenol) | | Descriptor: | 4,4'-(3-methylthiene-2,5-diyl)bis(3-chlorophenol), Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-15 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
7T8U
 
 | |
5DXR
 
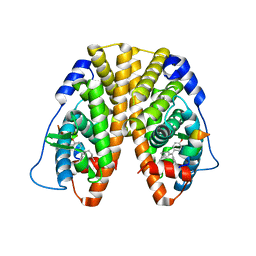 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 4,4'-{[(3R)-3-methylcyclohexylidene]methanediyl}diphenol | | Descriptor: | 4,4'-{[(3R)-3-methylcyclohexylidene]methanediyl}diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-23 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
7T0X
 
 | | Structure of the larger diameter PSMalpha3 nanotube | | Descriptor: | Phenol-soluble modulin PSM-alpha-3 | | Authors: | Kreutzberger, M.A, Wang, S, Beltran, L.C, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2021-11-30 | | Release date: | 2022-05-18 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Phenol-soluble modulins PSM alpha 3 and PSM beta 2 form nanotubes that are cross-alpha amyloids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SZZ
 
 | | Structure of the smaller diameter PSMalpha3 nanotubes | | Descriptor: | Phenol-soluble modulin PSM-alpha-3 | | Authors: | Beltran, L.C, Kreutzberger, M.A, Wang, S, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2021-11-29 | | Release date: | 2022-05-18 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Phenol-soluble modulins PSM alpha 3 and PSM beta 2 form nanotubes that are cross-alpha amyloids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5DWJ
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with a Resorcinyl 4-Fluoro-substituted Diaryl-imine analog 4-[(E)-[(4-fluorophenyl)imino](4-hydroxyphenyl)methyl]benzene-1,3-diol | | Descriptor: | 4-[(E)-[(4-fluorophenyl)imino](4-hydroxyphenyl)methyl]benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Wright, N.J, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-22 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DZI
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 4,4'-{[(3S)-3-(2-hydroxyethyl)cyclohexylidene]methanediyl}diphenol | | Descriptor: | 4,4'-{[(3S)-3-(2-hydroxyethyl)cyclohexylidene]methanediyl}diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-25 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DWI
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with a Resorcinyl 2-Chloro-substituted Diaryl-imine analog 4-[(E)-[(2-chlorophenyl)imino](4-hydroxyphenyl)methyl]benzene-1,3-diol | | Descriptor: | 4-[(E)-[(2-chlorophenyl)imino](4-hydroxyphenyl)methyl]benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Wright, N.J, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-22 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
