2F4M
 
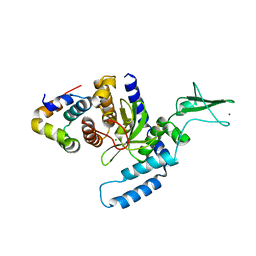 | | The Mouse PNGase-HR23 Complex Reveals a Complete Remodulation of the Protein-Protein Interface Compared to its Yeast Orthologs | | Descriptor: | CHLORIDE ION, UV excision repair protein RAD23 homolog B, ZINC ION, ... | | Authors: | Zhao, G, Zhou, X, Wang, L, Kisker, C, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2005-11-23 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the mouse peptide N-glycanase-HR23 complex suggests co-evolution of the endoplasmic reticulum-associated degradation and DNA repair pathways.
J.Biol.Chem., 281, 2006
|
|
1NYK
 
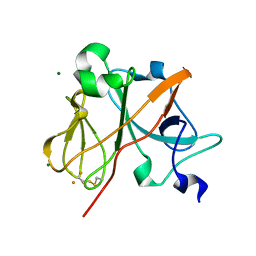 | | Crystal Structure of the Rieske protein from Thermus thermophilus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, MAGNESIUM ION, Rieske iron-sulfur protein | | Authors: | Hunsicker-Wang, L.M, Heine, A, Chen, Y, Luna, E.P, Todaro, T, Zhang, Y.M, Williams, P.A, McRee, D.E, Hirst, J, Stout, C.D, Fee, J.A. | | Deposit date: | 2003-02-12 | | Release date: | 2003-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | High resolution structure of the soluble, respiratory-type Rieske protein from Thermus thermophilus: Analysis and Comparison
Biochemistry, 42, 2003
|
|
2G9G
 
 | | Crystal structure of His-tagged mouse PNGase C-terminal domain | | Descriptor: | GLYCEROL, SULFATE ION, peptide N-glycanase | | Authors: | Zhou, X, Zhao, G, Wang, L, Li, G, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2006-03-06 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical studies of the C-terminal domain of mouse peptide-N-glycanase identify it as a mannose-binding module.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3TG5
 
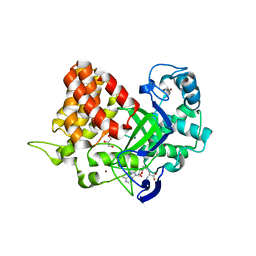 | | Structure of SMYD2 in complex with p53 and SAH | | Descriptor: | Cellular tumor antigen p53, GLYCEROL, N-lysine methyltransferase SMYD2, ... | | Authors: | Zhao, K, Wang, L. | | Deposit date: | 2011-08-17 | | Release date: | 2011-08-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of human SMYD2 reveals the basis of p53 tumor suppressor methylation
J.Biol.Chem., 2011
|
|
1OS5
 
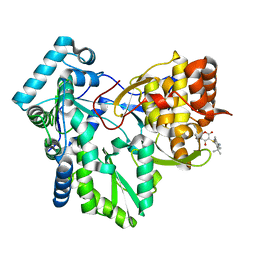 | | Crystal structure of HCV NS5B RNA polymerase complexed with a novel non-competitive inhibitor. | | Descriptor: | 3-(4-AMINO-2-TERT-BUTYL-5-METHYL-PHENYLSULFANYL)-6-CYCLOPENTYL-4-HYDROXY-6-[2-(4-HYDROXY-PHENYL)-ETHYL]-5,6-DIHYDRO-PYRAN-2-ONE, Hepatitis C virus NS5B RNA polymerase | | Authors: | Love, R.A, Parge, H.E, Yu, X, Hickey, M.J, Diehl, W, Gao, J, Wriggers, H, Ekker, A, Wang, L, Thomson, J.A, Dragovich, P.S, Fuhrman, S.A. | | Deposit date: | 2003-03-18 | | Release date: | 2004-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic identification of a noncompetitive inhibitor binding site on the hepatitis C virus NS5B RNA polymerase enzyme.
J.Virol., 77, 2003
|
|
1PC4
 
 | | Crystal Structure of the P50A mutant of ferredoxin I at 1.65 A Resolution | | Descriptor: | FE3-S4 CLUSTER, Ferredoxin I, IRON/SULFUR CLUSTER | | Authors: | Camba, R, Jung, Y.S, Chen, K, Hunsicker-Wang, L.M, Burgess, B.K, Stout, C.D, Hirst, J, Armstrong, F.A. | | Deposit date: | 2003-05-15 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanisms of redox-coupled proton transfer in proteins: role of the proximal proline in reactions of the [3Fe-4S] cluster in Azotobacter vinelandii ferredoxin I
Biochemistry, 42, 2003
|
|
1PC5
 
 | | Crystal Structure of the P50G Mutant of Ferredoxin I at 1.8 A Resolution | | Descriptor: | FE3-S4 CLUSTER, Ferredoxin I, IRON/SULFUR CLUSTER | | Authors: | Camba, R, Jung, Y.S, Chen, K, Hunsicker-Wang, L.M, Burgess, B.K, Stout, C.D, Hirst, J, Armstrong, F.A. | | Deposit date: | 2003-05-15 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanisms of redox-coupled proton transfer in proteins: role of the proximal proline in reactions of the [3Fe-4S] cluster in Azotobacter vinelandii ferredoxin I
Biochemistry, 42, 2003
|
|
5XVL
 
 | | Crystal structure of AL2 PAL domain | | Descriptor: | PHD finger protein ALFIN-LIKE 2, SULFATE ION | | Authors: | Peng, L, Wang, L.L, Huang, Y. | | Deposit date: | 2017-06-28 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Structural Analysis of the Arabidopsis AL2-PAL and PRC1 Complex Provides Mechanistic Insight into Active-to-Repressive Chromatin State Switch
J. Mol. Biol., 430, 2018
|
|
5XVJ
 
 | | Crystal structure of AL7 PAL domain | | Descriptor: | PHD finger protein ALFIN-LIKE 7, SULFATE ION | | Authors: | Peng, L, Wang, L.L, Huang, Y. | | Deposit date: | 2017-06-28 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Analysis of the Arabidopsis AL2-PAL and PRC1 Complex Provides Mechanistic Insight into Active-to-Repressive Chromatin State Switch
J. Mol. Biol., 430, 2018
|
|
5Y21
 
 | |
7E6T
 
 | | Structural insights into the activation of human calcium-sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYCLOMETHYLTRYPTOPHAN, ... | | Authors: | Geng, Y, Chen, X.C, Wang, L, Cui, Q.Q, Ding, Z.Y, Han, L, Kou, Y.J, Zhang, W.Q, Wang, H.N, Jia, X.M, Dai, M, Shi, Z.Z, Li, Y.Y, Li, X.Y. | | Deposit date: | 2021-02-24 | | Release date: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the activation of human calcium-sensing receptor.
Elife, 10, 2021
|
|
7E5R
 
 | | SARS-CoV-2 S trimer with three-antibody cocktail complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FC05 heavy chain, FC05 light chain, ... | | Authors: | Sun, Y, Wang, L, Wang, N, Feng, R, Wang, X. | | Deposit date: | 2021-02-20 | | Release date: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure-based development of three- and four-antibody cocktails against SARS-CoV-2 via multiple mechanisms.
Cell Res., 31, 2021
|
|
7E5S
 
 | | SARS-CoV-2 S trimer with four-antibody cocktail complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FC05 heavy chain, FC05 light chain, ... | | Authors: | Sun, Y, Wang, L, Wang, N, Feng, R, Wang, X. | | Deposit date: | 2021-02-20 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure-based development of three- and four-antibody cocktails against SARS-CoV-2 via multiple mechanisms.
Cell Res., 31, 2021
|
|
5G0F
 
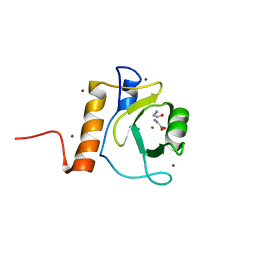 | | Crystal structure of Danio rerio HDAC6 ZnF-UBP domain | | Descriptor: | GLYCINE, HDAC6, NICKEL (II) ION, ... | | Authors: | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
5G0J
 
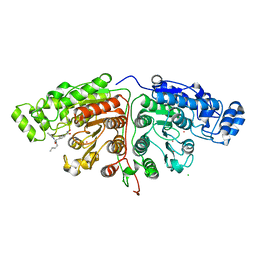 | | Crystal structure of Danio rerio HDAC6 CD1 and CD2 (linker intact) in complex with Nexturastat A | | Descriptor: | CHLORIDE ION, HDAC6, NEXTURASTAT A, ... | | Authors: | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
5G0G
 
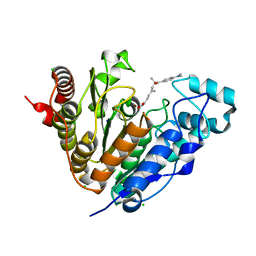 | | Crystal structure of Danio rerio HDAC6 CD1 in complex with trichostatin A | | Descriptor: | CHLORIDE ION, HDAC6, SODIUM ION, ... | | Authors: | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
5GI4
 
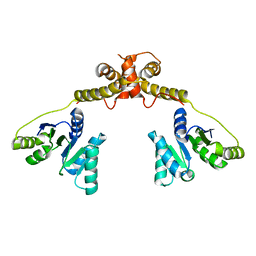 | | DEAD-box RNA helicase | | Descriptor: | ATP-dependent RNA helicase DeaD | | Authors: | Xu, L, Wang, L, Li, F, Wu, L, Shi, Y. | | Deposit date: | 2016-06-22 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Insights into the Structure of Dimeric RNA Helicase CsdA and Indispensable Role of Its C-Terminal Regions.
Structure, 25, 2017
|
|
7ENQ
 
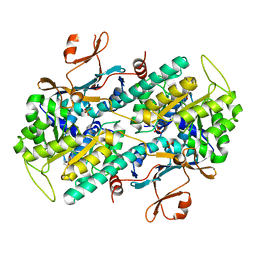 | | Crystal structure of human NAMPT in complex with compound NAT | | Descriptor: | 2-(2-~{tert}-butylphenoxy)-~{N}-(4-hydroxyphenyl)ethanamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Wang, G, Wu, C, Liu, M, Yao, H, Li, C, Wang, L, Tang, Y. | | Deposit date: | 2021-04-19 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.204966 Å) | | Cite: | Discovery of small-molecule activators of nicotinamide phosphoribosyltransferase (NAMPT) and their preclinical neuroprotective activity.
Cell Res., 32, 2022
|
|
1S68
 
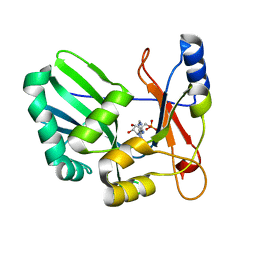 | | Structure and Mechanism of RNA Ligase | | Descriptor: | ADENOSINE MONOPHOSPHATE, RNA Ligase 2 | | Authors: | Ho, C.K, Wang, L.K, Lima, C.D, Shuman, S. | | Deposit date: | 2004-01-22 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of RNA ligase.
Structure, 12, 2004
|
|
4AG5
 
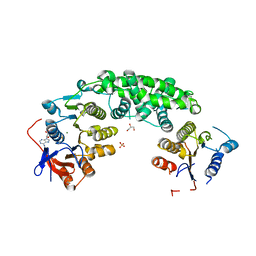 | | Structure of VirB4 of Thermoanaerobacter pseudethanolicus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wallden, K, Williams, R, Yan, J, Lian, P.W, Wang, L, Thalassinos, K, Orlova, E.V, Waksman, G. | | Deposit date: | 2012-01-24 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the Virb4 ATPase, Alone and Bound to the Core Complex of a Type Iv Secretion System.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TC6
 
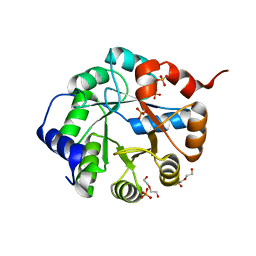 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR63. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Indole-3-glycerol phosphate synthase, PHOSPHATE ION | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Exploration of alternate catalytic mechanisms and optimization strategies for retroaldolase design.
J.Mol.Biol., 426, 2014
|
|
3TC7
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR62. | | Descriptor: | ACETIC ACID, Indole-3-glycerol phosphate synthase, PHOSPHATE ION | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Exploration of alternate catalytic mechanisms and optimization strategies for retroaldolase design.
J.Mol.Biol., 426, 2014
|
|
2PVE
 
 | | NMR and X-ray Analysis of Structural Additivity in Metal Binding Site-Swapped Hybrids of Rubredoxin | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Rubredoxin, ... | | Authors: | LeMaster, D.M, Anderson, J.S, Wang, L, Guo, Y, Li, H, Hernandez, G. | | Deposit date: | 2007-05-09 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.79 Å) | | Cite: | NMR and X-ray analysis of structural additivity in metal binding site-swapped hybrids of rubredoxin.
Bmc Struct.Biol., 7, 2007
|
|
5GJU
 
 | | DEAD-box RNA helicase | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-dependent RNA helicase DeaD | | Authors: | Xu, L, Li, F, Wang, L, Shi, Y. | | Deposit date: | 2016-07-02 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into the Structure of Dimeric RNA Helicase CsdA and Indispensable Role of Its C-Terminal Regions.
Structure, 25, 2017
|
|
3E84
 
 | | Structure-function Analysis of 2-Keto-3-deoxy-D-glycero-D-galacto-nononate-9-phosphate (KDN) Phosphatase Defines a New Clad Within the Type C0 HAD Subfamily | | Descriptor: | 1,2-ETHANEDIOL, Acylneuraminate cytidylyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lu, Z, Wang, L, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2008-08-19 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Function Analysis of 2-Keto-3-deoxy-D-glycero-D-galactonononate-9-phosphate Phosphatase Defines Specificity Elements in Type C0 Haloalkanoate Dehalogenase Family Members.
J.Biol.Chem., 284, 2009
|
|
