4DU1
 
 | | RB69 DNA Polymerase Ternary Complex with dATP Opposite dT | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Probing minor groove hydrogen bonding interactions between RB69 DNA polymerase and DNA.
Biochemistry, 51, 2012
|
|
4DTS
 
 | | RB69 DNA Polymerase Ternary Complex with dCTP Opposite an Abasic Site and ddC/dG as the Penultimate Base-pair | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Contribution of Partial Charge Interactions and Base Stacking to the Efficiency of Primer Extension at and beyond Abasic Sites in DNA.
Biochemistry, 51, 2012
|
|
4DTN
 
 | | RB69 DNA Polymerase Ternary Complex with dATP Opposite an Abasic Site and ddA/dT as the Penultimate Base-pair | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Contribution of Partial Charge Interactions and Base Stacking to the Efficiency of Primer Extension at and beyond Abasic Sites in DNA.
Biochemistry, 51, 2012
|
|
4DTU
 
 | | RB69 DNA Polymerase Ternary Complex with dGTP Opposite an Abasic Site and ddC/dG as the Penultimate Base-pair | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Contribution of Partial Charge Interactions and Base Stacking to the Efficiency of Primer Extension at and beyond Abasic Sites in DNA.
Biochemistry, 51, 2012
|
|
4EAL
 
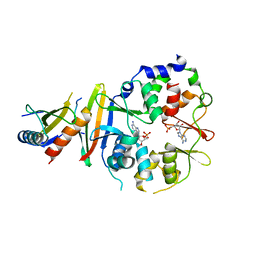 | | Co-crystal of AMPK core with ATP soaked with AMP | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Chen, L, Wang, J, Zhang, Y.-Y, Yan, S.F, Neumann, D, Schlattner, U, Wang, Z.-X, Wu, J.-W. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | AMP-activated protein kinase undergoes nucleotide-dependent conformational changes
Nat.Struct.Mol.Biol., 19, 2012
|
|
6M6E
 
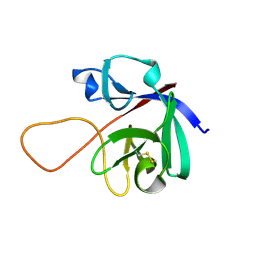 | |
6M6F
 
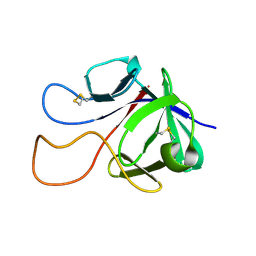 | |
1TF7
 
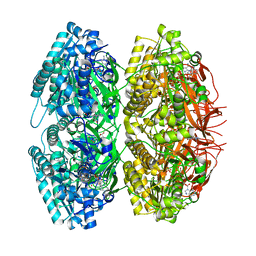 | | Crystal Structure of Circadian Clock Protein KaiC | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, KaiC | | Authors: | Pattanayek, R, Wang, J, Mori, T, Xu, Y, Johnson, C.H, Egli, M. | | Deposit date: | 2004-05-26 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Visualizing a Circadian Clock Protein; Crystal Structure of KaiC and Functional Insights
Mol.Cell, 15, 2004
|
|
4ZYN
 
 | | Crystal Structure of Parkin E3 ubiquitin ligase (linker deletion; delta 86-130) | | Descriptor: | E3 ubiquitin-protein ligase parkin, SULFATE ION, ZINC ION | | Authors: | Lilov, A, Sauve, V, Trempe, J.F, Rodionov, D, Wang, J, Gehring, K. | | Deposit date: | 2015-05-21 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | A Ubl/ubiquitin switch in the activation of Parkin.
Embo J., 34, 2015
|
|
5Z10
 
 | | Structure of the mechanosensitive Piezo1 channel | | Descriptor: | Piezo-type mechanosensitive ion channel component 1 | | Authors: | Zhao, Q, Zhou, H, Chi, S, Wang, Y, Wang, J, Geng, J, Wu, K, Liu, W, Zhang, T, Dong, M.-Q, Wang, J, Li, X, Xiao, B. | | Deposit date: | 2017-12-22 | | Release date: | 2018-01-31 | | Last modified: | 2020-01-29 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structure and mechanogating mechanism of the Piezo1 channel.
Nature, 554, 2018
|
|
5XWY
 
 | | Electron cryo-microscopy structure of LbuCas13a-crRNA binary complex | | Descriptor: | A type VI-A CRISPR-Cas RNA-guided RNA ribonuclease, Cas13a, RNA (59-MER) | | Authors: | Zhang, X, Wang, Y, Ma, J, Liu, L, Li, X, Li, Z, You, L, Wang, J, Wang, M. | | Deposit date: | 2017-06-30 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a.
Cell, 170, 2017
|
|
4EAG
 
 | | Co-crystal structure of an chimeric AMPK core with ATP | | Descriptor: | 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, L, Wang, J, Zhang, Y.-Y, Yan, S.F, Neumann, D, Schlattner, U, Wang, Z.-X, Wu, J.-W. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | AMP-activated protein kinase undergoes nucleotide-dependent conformational changes
Nat.Struct.Mol.Biol., 19, 2012
|
|
1I5T
 
 | | SOLUTION STRUCTURE OF CYANOFERRICYTOCHROME C | | Descriptor: | CYANIDE ION, CYTOCHROME C, HEME C | | Authors: | Yao, Y, Qian, C, Ye, K, Wang, J, Tang, W. | | Deposit date: | 2001-02-28 | | Release date: | 2001-03-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cyanoferricytochrome c: ligand-controlled conformational flexibility and electronic structure of the heme moiety.
J.Biol.Inorg.Chem., 7, 2002
|
|
1R8A
 
 | | Crystal Structures of an Archaeal Class I CCA-Adding Enzyme and Its Nucleotide Complexes | | Descriptor: | MANGANESE (II) ION, SODIUM ION, tRNA nucleotidyltransferase | | Authors: | Xiong, Y, Li, F, Wang, J, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes
Mol.Cell, 12, 2003
|
|
4N77
 
 | |
1S03
 
 | |
1R8B
 
 | | Crystal Structures of an Archaeal Class I CCA-Adding Enzyme and Its Nucleotide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Xiong, Y, Li, F, Wang, J, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes
Mol.Cell, 12, 2003
|
|
3KVW
 
 | | Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2) in complex with an indirubin ligand | | Descriptor: | (2Z,3E)-7'-bromo-3-(hydroxyimino)-2'-oxo-1,1',2',3-tetrahydro-2,3'-biindole-5-carboxylic acid, CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Filippakopoulos, P, Myrianthopoulos, V, Kritsanida, M, Magiatis, P, Skaltsounis, A.L, Soundararajan, M, Krojer, T, Gileadi, O, Hapka, E, Fedorov, O, Berridge, G, Wang, J, Shrestha, L, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Mikros, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-30 | | Release date: | 2010-01-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2) in complex with an indirubin ligand
To be Published
|
|
4EAI
 
 | | Co-crystal structure of an AMPK core with AMP | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Chen, L, Wang, J, Zhang, Y.-Y, Yan, S.F, Neumann, D, Schlattner, U, Wang, Z.-X, Wu, J.-W. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | AMP-activated protein kinase undergoes nucleotide-dependent conformational changes
Nat.Struct.Mol.Biol., 19, 2012
|
|
4EAK
 
 | | Co-crystal structure of an AMPK core with ATP | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Chen, L, Wang, J, Zhang, Y.-Y, Yan, S.F, Neumann, D, Schlattner, U, Wang, Z.-X, Wu, J.-W. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | AMP-activated protein kinase undergoes nucleotide-dependent conformational changes
Nat.Struct.Mol.Biol., 19, 2012
|
|
1R89
 
 | | Crystal Structures of an Archaeal Class I CCA-Adding Enzyme and Its Nucleotide Complexes | | Descriptor: | CHLORIDE ION, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xiong, Y, Li, F, Wang, J, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes
Mol.Cell, 12, 2003
|
|
1R8C
 
 | | Crystal Structures of an Archaeal Class I CCA-Adding Enzyme and Its Nucleotide | | Descriptor: | MANGANESE (II) ION, SODIUM ION, URIDINE 5'-TRIPHOSPHATE, ... | | Authors: | Xiong, Y, Li, F, Wang, J, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes
Mol.Cell, 12, 2003
|
|
7W0D
 
 | | Dicer2-LoqsPD-dsRNA complex at mid-translocation state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dicer-2, isoform A, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
7W0C
 
 | | Dicer2-Loqs-PD-dsRNA complex at early-translocation state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dicer-2, isoform A, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
7W0E
 
 | | dmDicer2-LoqsPD-dsRNA Active-dicing status | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dicer-2, isoform A, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
