7YKD
 
 | | Cryo-EM structure of the human chemerin receptor 1 complex with the C-terminal nonapeptide of chemerin | | Descriptor: | CHOLESTEROL, Chemerin-like receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Chen, G, Liao, Q, Ye, R.D, Wang, J. | | Deposit date: | 2022-07-22 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-EM structure of the human chemerin receptor 1-Gi protein complex bound to the C-terminal nonapeptide of chemerin.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3SUN
 
 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dTTP Opposite 2AP (AT rich sequence) | | Descriptor: | 5'-D(*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*(2DA))-3', 5'-D(P*CP*(2PR)P*TP*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*G)-3', CALCIUM ION, ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-11 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure of the 2-Aminopurine-Cytosine Base Pair Formed in the Polymerase Active Site of the RB69 Y567A-DNA Polymerase.
Biochemistry, 50, 2011
|
|
3SQ2
 
 | | RB69 DNA Polymerase Ternary Complex with dTTP Opposite 2AP (AT rich sequence) | | Descriptor: | 5'-D(*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*(2DA))-3', 5'-D(P*CP*(2PR)P*TP*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*G)-3', CALCIUM ION, ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-04 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the 2-Aminopurine-Cytosine Base Pair Formed in the Polymerase Active Site of the RB69 Y567A-DNA Polymerase.
Biochemistry, 50, 2011
|
|
3SUO
 
 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dTTP Opposite 2AP (GC rich sequence) | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*GP*GP*CP*GP*GP*CP*GP*(2DA))-3', 5'-D(P*CP*(2PR)P*TP*CP*GP*CP*CP*GP*CP*CP*GP*CP*GP*CP*GP*G)-3', CALCIUM ION, ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-11 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure of the 2-Aminopurine-Cytosine Base Pair Formed in the Polymerase Active Site of the RB69 Y567A-DNA Polymerase.
Biochemistry, 50, 2011
|
|
3SQ4
 
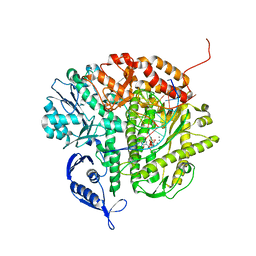 | | RB69 DNA Polymerase Ternary Complex with dTTP Opposite 2AP (GC rich sequence) | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*GP*GP*CP*GP*GP*CP*GP*(2DA))-3', 5'-D(P*CP*(2PR)P*TP*CP*GP*CP*CP*GP*CP*CP*GP*CP*GP*CP*GP*G)-3', CALCIUM ION, ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-04 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure of the 2-Aminopurine-Cytosine Base Pair Formed in the Polymerase Active Site of the RB69 Y567A-DNA Polymerase.
Biochemistry, 50, 2011
|
|
3SUQ
 
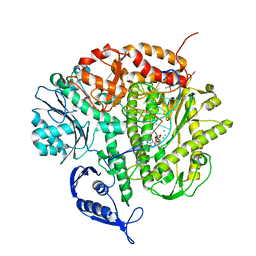 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dCTP Opposite 2AP (AT rich sequence) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*(2DA))-3', 5'-D(P*CP*(2PR)P*TP*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*G)-3', ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-11 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of the 2-Aminopurine-Cytosine Base Pair Formed in the Polymerase Active Site of the RB69 Y567A-DNA Polymerase.
Biochemistry, 50, 2011
|
|
3SUP
 
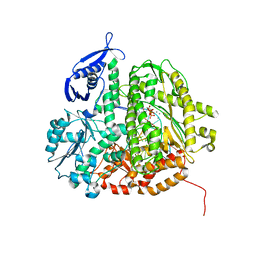 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dCTP Opposite 2AP (GC rich sequence) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*GP*CP*GP*CP*GP*GP*CP*GP*GP*CP*GP*(2DA))-3', 5'-D(P*CP*(2PR)P*TP*CP*GP*CP*CP*GP*CP*CP*GP*CP*GP*CP*GP*G)-3', ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-11 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure of the 2-Aminopurine-Cytosine Base Pair Formed in the Polymerase Active Site of the RB69 Y567A-DNA Polymerase.
Biochemistry, 50, 2011
|
|
7JI2
 
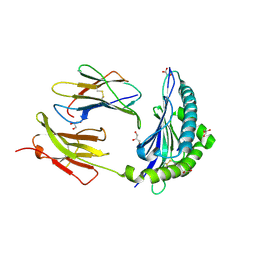 | | Crystal Structure of H2-Kb in complex with a OVA mutant peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Li, X, Mallis, R.J, Mizsei, R, Tan, K, Reinherz, E.L, Wang, J. | | Deposit date: | 2020-07-22 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pre-T cell receptors topologically sample self-ligands during thymocyte beta-selection.
Science, 371, 2021
|
|
1FWS
 
 | | AQUIFEX AEOLICUS KDO8P SYNTHASE IN COMPLEX WITH PEP AND CADMIUM | | Descriptor: | 2-DEHYDRO-3-DEOXYPHOSPHOOCTONATE ALDOLASE, CADMIUM ION, PHOSPHATE ION, ... | | Authors: | Duewel, H.S, Radaev, S, Wang, J, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2000-09-24 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Electronic structure of the metal center in the Cd(2+), Zn(2+), and Cu(2+) substituted forms of KDO8P synthase: implications for catalysis.
Biochemistry, 48, 2009
|
|
8GTZ
 
 | |
1FWW
 
 | | AQUIFEX AEOLICUS KDO8P SYNTHASE IN COMPLEX WITH PEP, A5P AND CADMIUM | | Descriptor: | 2-DEHYDRO-3-DEOXYPHOSPHOOCTONATE ALDOLASE, ARABINOSE-5-PHOSPHATE, CADMIUM ION, ... | | Authors: | Duewel, H.S, Radaev, S, Wang, J, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2000-09-24 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Electronic structure of the metal center in the Cd(2+), Zn(2+), and Cu(2+) substituted forms of KDO8P synthase: implications for catalysis.
Biochemistry, 48, 2009
|
|
4AF3
 
 | | Human Aurora B Kinase in complex with INCENP and VX-680 | | Descriptor: | AURORA KINASE B, CYCLOPROPANECARBOXYLIC ACID {4-[4-(4-METHYL-PIPERAZIN-1-YL)-6-(5-METHYL-2H-PYRAZOL-3-YLAMINO)-PYRIMIDIN-2-YLSULFANYL]-PHENYL}-AMIDE, INNER CENTROMERE PROTEIN | | Authors: | Elkins, J.M, Vollmar, M, Wang, J, Picaud, S, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Knapp, S. | | Deposit date: | 2012-01-16 | | Release date: | 2012-04-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of Human Aurora B in Complex with Incenp and Vx-680.
J.Med.Chem., 55, 2012
|
|
4J2B
 
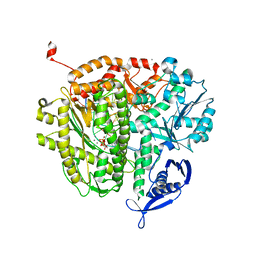 | | RB69 DNA Polymerase L415G Ternary Complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*T)-3'), ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2013-02-04 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Alteration in the cavity size adjacent to the active site of RB69 DNA polymerase changes its conformational dynamics.
Nucleic Acids Res., 41, 2013
|
|
4J2A
 
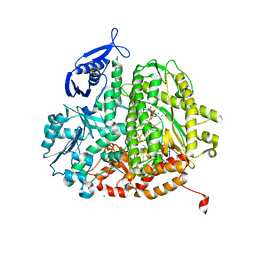 | | RB69 DNA Polymerase L415A Ternary Complex | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*C)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3'), ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2013-02-04 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alteration in the cavity size adjacent to the active site of RB69 DNA polymerase changes its conformational dynamics.
Nucleic Acids Res., 41, 2013
|
|
4J2E
 
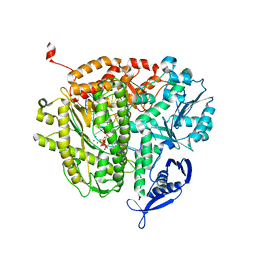 | | RB69 DNA Polymerase L415M Ternary Complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*G)-3'), ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2013-02-04 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Alteration in the cavity size adjacent to the active site of RB69 DNA polymerase changes its conformational dynamics.
Nucleic Acids Res., 41, 2013
|
|
2LUN
 
 | |
8KA0
 
 | | Crystal structure of Vibrio vulnificus RID-dependent transforming NADase domain (RDTND)/calmodulin-binding domain of Rho inactivation domain (RID-CBD) complexed with Ca2+-bound calmodulin and a nicotinamide adenine dinucleotide (NAD+) | | Descriptor: | CALCIUM ION, Calmodulin-2, GLYCEROL, ... | | Authors: | Lee, Y, Choi, S, Hwang, J, Kim, M.H. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Dissemination of pathogenic bacteria is reinforced by a MARTX toxin effector duet.
Nat Commun, 15, 2024
|
|
8KA2
 
 | |
8K9Z
 
 | | Crystal structure of Vibrio vulnificus RID-dependent transforming NADase domain (RDTND)/calmodulin-binding domain of Rho inactivation domain (RID-CBD) complexed with Ca2+-bound calmodulin | | Descriptor: | CALCIUM ION, Calmodulin-2, RDTND-RID CBD | | Authors: | Lee, Y, Choi, S, Hwang, J, Kim, M.H. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dissemination of pathogenic bacteria is reinforced by a MARTX toxin effector duet.
Nat Commun, 15, 2024
|
|
8KA1
 
 | | Crystal structure of Vibrio vulnificus RID-dependent transforming NADase domain (RDTND)/calmodulin-binding domain of Rho inactivation domain (RID-CBD) complexed with Ca2+-free calmodulin | | Descriptor: | Calmodulin-2, MAGNESIUM ION, RDTND-RID CBD | | Authors: | Lee, Y, Choi, S, Hwang, J, Kim, M.H. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Dissemination of pathogenic bacteria is reinforced by a MARTX toxin effector duet.
Nat Commun, 15, 2024
|
|
5WDH
 
 | | Motor domain of human kinesin family member C1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIFC1, MAGNESIUM ION, ... | | Authors: | Zhu, H, Tempel, W, He, H, Shen, Y, Wang, J, Brothers, G, Landry, R, Arrowsmith, C.H, Edwards, A.M, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-07-05 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.248 Å) | | Cite: | Structural basis of small molecule ATPase inhibition of a human mitotic kinesin motor protein.
Sci Rep, 7, 2017
|
|
3KZ9
 
 | | Crystal structure of the master transcriptional regulator, SmcR, in Vibrio vulnificus provides insight into its DNA recognition mechanism | | Descriptor: | SULFATE ION, SmcR | | Authors: | Kim, M.H, Kim, Y, Choi, W.-C, Hwang, J. | | Deposit date: | 2009-12-08 | | Release date: | 2010-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of SmcR, a quorum-sensing master regulator of Vibrio vulnificus, provides insight into its regulation of transcription
J.Biol.Chem., 285, 2010
|
|
6KYV
 
 | | Crystal Structure of RIG-I and hairpin RNA with G-U wobble base pairs | | Descriptor: | Probable ATP-dependent RNA helicase DDX58, RNA (5'-R(*GP*GP*UP*AP*GP*AP*CP*GP*CP*UP*UP*CP*GP*GP*CP*GP*UP*UP*UP*GP*CP*C)-3'), ZINC ION | | Authors: | Kim, K.-H, Hwang, J, Kim, J.H, Son, K.-P, Jang, Y, Kim, M, Kang, S.-J, Lee, J.-O, Choi, B.-S. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biophysical properties of RIG-I bound to dsRNA with G-U wobble base pairs.
Rna Biol., 17, 2020
|
|
6L0Y
 
 | | Structure of dsRNA with G-U wobble base pairs | | Descriptor: | RNA (5'-R(*GP*GP*UP*AP*GP*AP*CP*GP*CP*UP*UP*CP*GP*GP*CP*GP*UP*UP*UP*GP*CP*C)-3') | | Authors: | Kim, K.-H, Hwang, J, Kim, J.H, Son, K.-P, Jang, Y, Kim, M, Kang, S.-J, Lee, J.-O, Choi, B.-S. | | Deposit date: | 2019-09-27 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of dsRNA with G-U wobble base pairs
To Be Published
|
|
7CS1
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis-Complex with Coenzyme A and Neomycin | | Descriptor: | Aminoglycoside 2'-N-acetyltransferase, COENZYME A, NEOMYCIN | | Authors: | Jeong, C.S, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.966 Å) | | Cite: | Structural and biochemical analyses of an aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis.
Sci Rep, 10, 2020
|
|
