6C69
 
 | | Structure of glycolipid aGSA[12,6P] in complex with mouse CD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
6C6E
 
 | | Structure of glycolipid aGSA[26,6P] in complex with mouse CD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
8IBX
 
 | | Structure of R2 with 3'UTR and DNA in unwinding state | | Descriptor: | 3'UTR, DNA (60-MER), Reverse transcriptase-like protein, ... | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
8IBW
 
 | | Structure of R2 with 3'UTR and DNA in binding state | | Descriptor: | 3'UTR, DNA (60-MER), Reverse transcriptase-like protein, ... | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
8IBZ
 
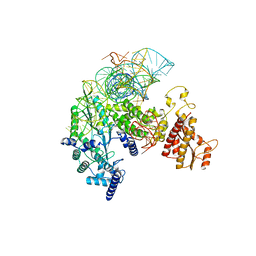 | | Structure of R2 with 5'ORF and 3'UTR | | Descriptor: | 5ORF-linker-3UTR, Reverse transcriptase-like protein, ZINC ION | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
8IBY
 
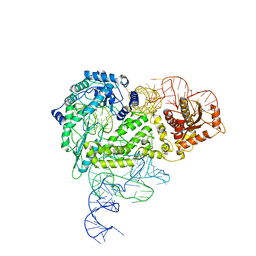 | | Structure of R2 with 5'ORF | | Descriptor: | 5'ORF RNA, Reverse transcriptase-like protein, ZINC ION | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
4EDK
 
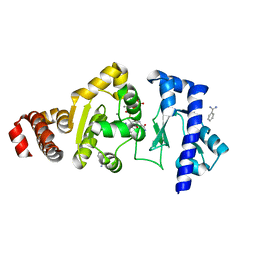 | | The structure of the S. aureus DnaG RNA Polymerase Domain bound to GTP and Manganese | | Descriptor: | BENZAMIDINE, DNA primase, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Rymer, R.U, Solorio, F.A, Chu, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-27 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
5FKP
 
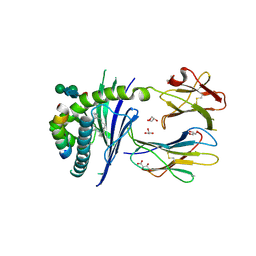 | | Crystal structure of the mouse CD1d in complex with the p99 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIGEN-PRESENTING GLYCOPROTEIN CD1D1, BETA 2 MICROGLOBULIN, ... | | Authors: | Girardi, E, Wang, J, Zajonc, D.M. | | Deposit date: | 2015-10-18 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of an Alpha-Helical Peptide and Lipopeptide Bound to the Non-Classical Mhc Class I Molecule Cd1D
J.Biol.Chem., 291, 2016
|
|
7LJY
 
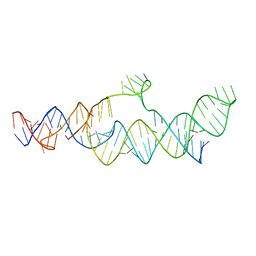 | | Cryo-EM structure of the B dENE construct complexed with a 28-mer poly(A) | | Descriptor: | B dENE construct, poly(A) | | Authors: | Torabi, S, Chen, Y, Zhang, K, Wang, J, DeGregorio, S, Vaidya, A, Su, Z, Pabit, S, Chiu, W, Pollack, L, Steitz, J. | | Deposit date: | 2021-02-01 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural analyses of an RNA stability element interacting with poly(A).
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4H42
 
 | | Synthesis of a Weak Basic uPA Inhibitor and Crystal Structure of Complex with uPA | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, N-[(2-amino-1,3-benzothiazol-6-yl)carbonyl]glycine, Urokinase-type plasminogen activator | | Authors: | Yu, H.-Y, Gao, D, Zhang, X, Jiang, L.-G, Hong, Z.-B, Yuan, C, Fang, X, Wang, J.-D, Huang, M.-D. | | Deposit date: | 2012-09-14 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Synthesis of a Weak Basic uPA Inhibitor and Crystal Structure of Complex with uPA
CHIN.J.STRUCT.CHEM., 32, 2013
|
|
6C6A
 
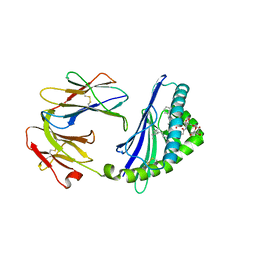 | | Structure of glycolipid aGSA[16,6P] in complex with mouse CD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
7LX0
 
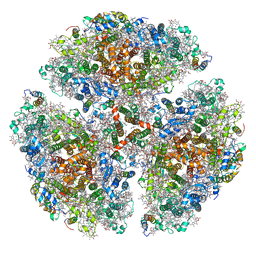 | | Quantitative assessment of chlorophyll types in cryo-EM maps of photosystem I acclimated to far-red light | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Gisriel, C.J, Wang, J. | | Deposit date: | 2021-03-02 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Quantitative assessment of chlorophyll types in cryo-EM maps of photosystem I acclimated to far-red light
BBA Adv, 1, 2021
|
|
2Z3A
 
 | | Crystal Structure of Bacillus Subtilis CodW, a non-canonical HslV-like peptidase with an impaired catalytic apparatus | | Descriptor: | ATP-dependent protease hslV | | Authors: | Rho, S.H, Park, H.H, Kang, G.B, Lim, Y.J, Kang, M.S, Lim, B.K, Seong, I.S, Chung, C.H, Wang, J, Eom, S.H. | | Deposit date: | 2007-06-03 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Bacillus subtilis CodW, a noncanonical HslV-like peptidase with an impaired catalytic apparatus
Proteins, 71, 2007
|
|
3PS4
 
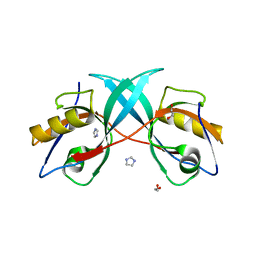 | | PDZ domain from Human microtubule-associated serine/threonine-protein kinase 1 | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, Microtubule-associated serine/threonine-protein kinase 1 | | Authors: | Ugochukwu, E, Wang, J, Krojer, T, Muniz, J.R.C, Sethi, R, Pike, A.C.W, Roos, A, Salah, E, Cocking, R, Savitsky, P, Doyle, D.A, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-30 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | PDZ domain from Human microtubule-associated serine/threonine-protein kinase 1
TO BE PUBLISHED
|
|
3Q5Y
 
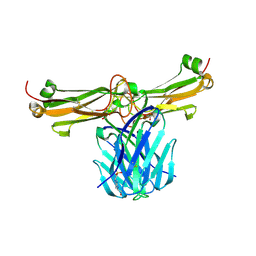 | | V beta/V beta homodimerization-based pre-TCR model suggested by TCR beta crystal structures | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Chen, Q, Zhang, H, Wang, J.-H. | | Deposit date: | 2010-12-30 | | Release date: | 2011-03-09 | | Last modified: | 2014-10-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A conserved hydrophobic patch on Vbeta domains revealed by TCRbeta chain crystal structures: implications for pre-TCR dimerization
Front Immunol, 2, 2011
|
|
4EDG
 
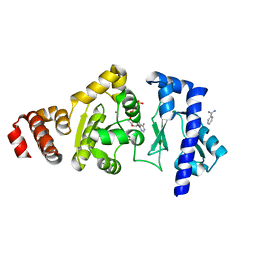 | | The structure of the S. aureus DnaG RNA Polymerase Domain bound to ATP and Manganese | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, BENZAMIDINE, DNA primase, ... | | Authors: | Rymer, R.U, Solorio, F.A, Chu, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-27 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
5A1Z
 
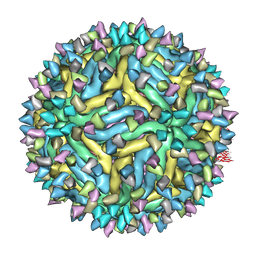 | | Cryo-EM structure of Dengue virus serotype 2 strain PVP94-07 complexed with human antibody 2D22 Fab at 37 degrees C | | Descriptor: | ANTIGEN-BINDING FRAGMENT OF HUMAN ANTIBODY 2D22 - HEAVY CHAIN, ANTIGEN-BINDING FRAGMENT OF HUMAN ANTIBODY 2D22 - LIGHT CHAIN, ENVELOPE PROTEIN, ... | | Authors: | Fibriansah, G, Ibarra, K.D, Ng, T.S, Smith, S.A, Tan, J.L, Lim, X.N, Ooi, J.S.G, Kostyuchenko, V.A, Wang, J, de Silva, A.M, Harris, E, Crowe Jr, J.E, Lok, S.M. | | Deposit date: | 2015-05-06 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers.
Science, 349, 2015
|
|
1FWW
 
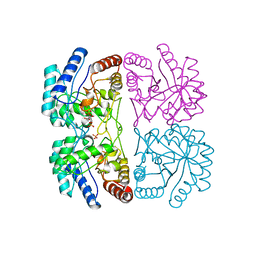 | | AQUIFEX AEOLICUS KDO8P SYNTHASE IN COMPLEX WITH PEP, A5P AND CADMIUM | | Descriptor: | 2-DEHYDRO-3-DEOXYPHOSPHOOCTONATE ALDOLASE, ARABINOSE-5-PHOSPHATE, CADMIUM ION, ... | | Authors: | Duewel, H.S, Radaev, S, Wang, J, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2000-09-24 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Electronic structure of the metal center in the Cd(2+), Zn(2+), and Cu(2+) substituted forms of KDO8P synthase: implications for catalysis.
Biochemistry, 48, 2009
|
|
4E2K
 
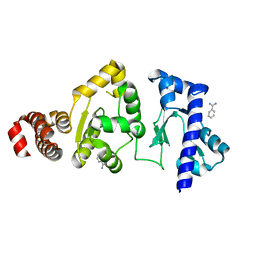 | | The structure of the S. aureus DnaG RNA Polymerase Domain | | Descriptor: | BENZAMIDINE, DNA primase | | Authors: | Rymer, R.U, Solorio, F.A, Clement, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-08 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
1FWS
 
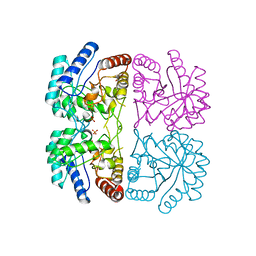 | | AQUIFEX AEOLICUS KDO8P SYNTHASE IN COMPLEX WITH PEP AND CADMIUM | | Descriptor: | 2-DEHYDRO-3-DEOXYPHOSPHOOCTONATE ALDOLASE, CADMIUM ION, PHOSPHATE ION, ... | | Authors: | Duewel, H.S, Radaev, S, Wang, J, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2000-09-24 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Electronic structure of the metal center in the Cd(2+), Zn(2+), and Cu(2+) substituted forms of KDO8P synthase: implications for catalysis.
Biochemistry, 48, 2009
|
|
3COO
 
 | |
4RNG
 
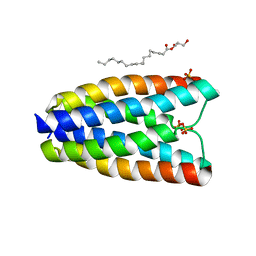 | | Crystal structure of a bacterial homologue of SWEET transporters | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MtN3/saliva family, SULFATE ION | | Authors: | Hu, Q, Wang, J, Yan, C, Yan, N. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a bacterial homologue of SWEET transporters.
Cell Res., 24, 2014
|
|
2LUA
 
 | | Solution structure of CXC domain of MSL2 | | Descriptor: | Protein male-specific lethal-2, ZINC ION | | Authors: | Feng, Y, Ye, K, Zheng, S, Wang, J. | | Deposit date: | 2012-06-09 | | Release date: | 2012-10-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of MSL2 CXC Domain Reveals an Unusual Zn(3)Cys(9) Cluster and Similarity to Pre-SET Domains of Histone Lysine Methyltransferases.
Plos One, 7, 2012
|
|
4FC6
 
 | | Studies on DCR shed new light on peroxisomal beta-oxidation: Crystal structure of the ternary complex of pDCR | | Descriptor: | HEXANOYL-COENZYME A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Peroxisomal 2,4-dienoyl-CoA reductase | | Authors: | Hua, T, Wu, D, Wang, J, Shaw, N, Liu, Z.-J. | | Deposit date: | 2012-05-24 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Studies of human 2,4-dienoyl CoA reductase shed new light on peroxisomal beta-oxidation of unsaturated fatty acids
J.Biol.Chem., 287, 2012
|
|
4FC7
 
 | | Studies on DCR shed new light on peroxisomal beta-oxidation: Crystal structure of the ternary complex of pDCR | | Descriptor: | COENZYME A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Peroxisomal 2,4-dienoyl-CoA reductase | | Authors: | Hua, T, Wu, D, Wang, J, Shaw, N, Liu, Z.-J. | | Deposit date: | 2012-05-24 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Studies of human 2,4-dienoyl CoA reductase shed new light on peroxisomal beta-oxidation of unsaturated fatty acids
J.Biol.Chem., 287, 2012
|
|
