8HW5
 
 | |
5XE0
 
 | | Crystal structure of EV-D68-3Dpol in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Genome polyprotein | | Authors: | Xie, W, Wang, C, Wang, Z, Li, Q, Wang, C. | | Deposit date: | 2017-03-30 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure and Thermostability Characterization of Enterovirus D68 3Dpol
J. Virol., 91, 2017
|
|
4DUM
 
 | | Co-crystal structure of eIF4E with inhibitor | | Descriptor: | (4-{7-[2-(4-chlorophenoxy)ethyl]-2-(methylamino)-6-oxo-6,7-dihydro-1H-purin-8-yl}phenyl)phosphonic acid, 1,2-ETHANEDIOL, Eukaryotic translation initiation factor 4E | | Authors: | Min, X, Johnstone, S, Walker, N, Wang, Z. | | Deposit date: | 2012-02-22 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure-Guided Design, Synthesis, and Evaluation of Guanine-Derived Inhibitors of the eIF4E mRNA-Cap Interaction.
J.Med.Chem., 55, 2012
|
|
4F2L
 
 | | Structure of a regulatory domain of AMPK | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, MAGNESIUM ION | | Authors: | Xin, F.J, Zhang, Y.Y, Wang, J, Wang, Z.X, Wu, J.W. | | Deposit date: | 2012-05-08 | | Release date: | 2013-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conserved regulatory elements in AMPK
Nature, 498, 2013
|
|
1A40
 
 | | PHOSPHATE-BINDING PROTEIN WITH ALA 197 REPLACED WITH TRP | | Descriptor: | PHOSPHATE ION, PHOSPHATE-BINDING PERIPLASMIC PROTEIN PRECURSOR | | Authors: | Ledivina, P.S, Wang, Z, Tsai, A, Koehl, E, Quiocho, F.A. | | Deposit date: | 1998-02-10 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dominant role of local dipolar interactions in phosphate binding to a receptor cleft with an electronegative charge surface: equilibrium, kinetic, and crystallographic studies.
Protein Sci., 7, 1998
|
|
4DT6
 
 | | Co-crystal structure of eIF4E with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 7-[2-(4-chlorophenoxy)ethyl]guanosine 5'-(dihydrogen phosphate), Eukaryotic translation initiation factor 4E, ... | | Authors: | Min, X, Johnstone, S, Walker, N, Wang, Z. | | Deposit date: | 2012-02-20 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Guided Design, Synthesis, and Evaluation of Guanine-Derived Inhibitors of the eIF4E mRNA-Cap Interaction.
J.Med.Chem., 55, 2012
|
|
5GNY
 
 | | The structure of WT Bgl6 | | Descriptor: | Beta-glucosidase, beta-D-glucopyranose | | Authors: | Xie, W, Pang, P, Cao, L.C, Liu, Y.H, Wang, Z. | | Deposit date: | 2016-07-25 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Structures of a glucose-tolerant beta-glucosidase provide insights into its mechanism.
J. Struct. Biol., 198, 2017
|
|
3P4Z
 
 | | Time-dependent and Protein-directed In Situ Growth of Gold Nanoparticles in a Single Crystal of Lysozyme | | Descriptor: | CHLORIDE ION, GOLD 3+ ION, GOLD ION, ... | | Authors: | Wei, H, Wang, Z, Zhang, J, House, S, Gao, Y.-G, Yang, L, Robinson, H, Tan, L.H, Xing, H, Hou, C, Robertson, I.M, Zuo, J.-M, Lu, Y. | | Deposit date: | 2010-10-07 | | Release date: | 2011-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-dependent, protein-directed growth of gold nanoparticles within a single crystal of lysozyme.
Nat Nanotechnol, 6, 2011
|
|
6UWD
 
 | | Crystal structure of Apo AtmM | | Descriptor: | ACETATE ION, D-glucose O-methyltransferase, MAGNESIUM ION | | Authors: | Alvarado, S.K, Wang, Z, Miller, M.D, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-11-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of Apo AtmM
To Be Published
|
|
3P64
 
 | | Time-dependent and Protein-directed In Situ Growth of Gold Nanoparticles in a Single Crystal of Lysozyme | | Descriptor: | CHLORIDE ION, GOLD 3+ ION, GOLD ION, ... | | Authors: | Wei, H, Wang, Z, Zhang, J, House, S, Gao, Y.-G, Yang, L, Robinson, H, Tan, L.H, Xing, H, Hou, C, Robertson, I.M, Zuo, J.-M, Lu, Y. | | Deposit date: | 2010-10-11 | | Release date: | 2011-02-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Time-dependent, protein-directed growth of gold nanoparticles within a single crystal of lysozyme.
Nat Nanotechnol, 6, 2011
|
|
5GNX
 
 | | The E171Q mutant structure of Bgl6 | | Descriptor: | Beta-glucosidase, GLYCEROL, PROPANOIC ACID, ... | | Authors: | Xie, W, Pang, P, Cao, L.C, Liu, Y.H, Wang, Z. | | Deposit date: | 2016-07-25 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of a glucose-tolerant beta-glucosidase provide insights into its mechanism.
J. Struct. Biol., 198, 2017
|
|
5GNZ
 
 | | The M3 mutant structure of Bgl6 | | Descriptor: | Beta-glucosidase, GLYCEROL, beta-D-glucopyranose | | Authors: | Xie, W, Pang, P, Cao, L.C, Liu, Y.H, Wang, Z. | | Deposit date: | 2016-07-25 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of a glucose-tolerant beta-glucosidase provide insights into its mechanism.
J. Struct. Biol., 198, 2017
|
|
1J4G
 
 | | crystal structure analysis of the trichosanthin delta C7 | | Descriptor: | CALCIUM ION, Trichosanthin delta C7 | | Authors: | Ding, Y, Too, H.M, Wang, Z.L, Liu, Y.W, Dong, Y.C, Shaw, P.C, Rao, Z.H. | | Deposit date: | 2001-09-30 | | Release date: | 2001-10-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure analysis of the trichosanthin delta C7
To be Published
|
|
3EQP
 
 | | Crystal Structure of Ack1 with compound T95 | | Descriptor: | Activated CDC42 kinase 1, CHLORIDE ION, N-(2,6-dimethylphenyl)-4-(2-ethoxyphenoxy)-2-({4-[4-(2-hydroxyethyl)piperazin-1-yl]phenyl}amino)pyrimidine-5-carboxamide | | Authors: | Liu, J, Wang, Z, Walker, N.P.C. | | Deposit date: | 2008-10-01 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and optimization of N3,N6-diaryl-1H-pyrazolo[3,4-d]pyrimidine-3,6-diamines as a novel class of ACK1 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3PY3
 
 | |
7WKH
 
 | | the grass carp IFNa | | Descriptor: | Interferon | | Authors: | Zou, J, WANG, J, Wang, Z. | | Deposit date: | 2022-01-09 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural and Functional Analyses of Type I IFNa Shed Light Into Its Interaction With Multiple Receptors in Fish.
Front Immunol, 13, 2022
|
|
8I4Z
 
 | | CalA3 with hydrolysis product | | Descriptor: | 11-oxidanylidene-11-(1~{H}-pyrrol-2-yl)undecanoic acid, Beta-ketoacyl-acyl-carrier-protein synthase I | | Authors: | Wang, J, Wang, Z. | | Deposit date: | 2023-01-21 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | C-N bond formation by a polyketide synthase.
Nat Commun, 14, 2023
|
|
7WP9
 
 | | SARS-CoV-2 Omicron Variant SPIKE trimer, all RBDs down | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
8I4Y
 
 | | CalA3 complex structure with amidation product | | Descriptor: | 11-oxidanylidene-11-(1~{H}-pyrrol-2-yl)undecanoic acid, 3-HYDROXYANTHRANILIC ACID, Beta-ketoacyl-acyl-carrier-protein synthase I | | Authors: | Wang, J, Wang, Z. | | Deposit date: | 2023-01-21 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | C-N bond formation by a polyketide synthase.
Nat Commun, 14, 2023
|
|
7WPB
 
 | | SARS-CoV-2 Omicron Variant RBD complexed with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPC
 
 | | The second RBD of SARS-CoV-2 Omicron Variant in complexed with RBD-ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
5FDX
 
 | | Structure of DDR1 receptor tyrosine kinase in complex with D2164 inhibitor at 2.65 Angstroms resolution. | | Descriptor: | 1,2-ETHANEDIOL, 3-[(4-methylpiperazin-1-yl)methyl]-~{N}-[(4~{R})-4-methyl-2-pyrimidin-5-yl-3,4-dihydro-1~{H}-isoquinolin-7-yl]-5-(trifluoromethyl)benzamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bartual, S.G, Pinkas, D.M, Wang, Z, Ding, K, Mahajan, P, Kupinska, K, Mukhopadhyay, S, Strain-Damerell, C, Chalk, R, Borkowska, O, Talon, R, Kopec, J, Williams, E, Tallant, C, Chaikuad, A, Sorell, F, Newman, J, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of DDR1 receptor tyrosine kinase in complex with D2164 inhibitor at 2.65 Angstroms resolution.
To Be Published
|
|
5X6G
 
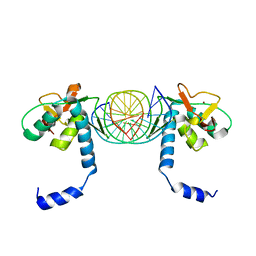 | | Crystal Structure of SMAD5-MH1/palindromic SBE DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*CP*AP*GP*TP*CP*TP*AP*GP*AP*CP*AP*TP*A)-3'), DNA (5'-D(P*GP*TP*AP*TP*GP*TP*CP*TP*AP*GP*AP*CP*TP*GP*A)-3'), Mothers against decapentaplegic homolog 5, ... | | Authors: | Chai, N, Wang, J, Wang, Z.X, Wu, J.W. | | Deposit date: | 2017-02-21 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis for the Smad5 MH1 domain to recognize different DNA sequences.
Nucleic Acids Res., 43, 2015
|
|
5X6M
 
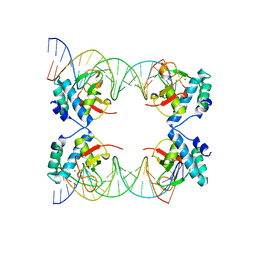 | | Crystal Structure of SMAD5-MH1 in complex with a composite DNA sequence | | Descriptor: | DNA (5'-D(P*AP*TP*CP*AP*GP*AP*CP*TP*GP*CP*CP*GP*GP*CP*AP*GP*TP*CP*TP*AP*TP*A)-3'), DNA (5'-D(P*TP*TP*AP*TP*AP*GP*AP*CP*TP*GP*CP*CP*GP*GP*CP*AP*GP*TP*CP*TP*GP*A)-3'), Mothers against decapentaplegic homolog 5, ... | | Authors: | Chai, N, Wang, J, Wang, Z.X, Wu, J.W. | | Deposit date: | 2017-02-22 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the Smad5 MH1 domain to recognize different DNA sequences.
Nucleic Acids Res., 43, 2015
|
|
6O3V
 
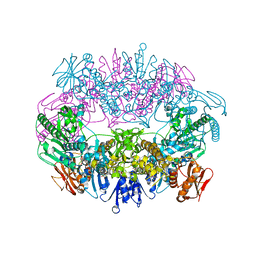 | | Crystal structure for RVA-VP3 | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-MONOPHOSPHATE, Protein VP3, ... | | Authors: | Kumar, D, Yu, X, Wang, Z, Hu, L, Prasad, V. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | 2.7 angstrom cryo-EM structure of rotavirus core protein VP3, a unique capping machine with a helicase activity.
Sci Adv, 6, 2020
|
|
