2Z1Q
 
 | |
6GT3
 
 | | Crystal Structure of the A2A-StaR2-bRIL562 in complex with AZD4635 at 2.0A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 6-(2-chloranyl-6-methyl-pyridin-4-yl)-5-(4-fluorophenyl)-1,2,4-triazin-3-amine, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, ... | | Authors: | Borodovsky, A, Wang, Y, Deng, N, Ye, M, Stephen, T.L, Goodwin, K, Goodwin, R, Strittmatter, N, Shaw, J, Sachsenmeier, K, Clarke, J.D, Hay, C, Reimer, C, Andrews, S.P, Brown, G.A, Congreve, M, Cheng, R.K.Y, Dore, A.S, Mason, J.S, Marshall, F.H, Weir, M.P, Lyne, P, Woessner, R. | | Deposit date: | 2018-06-15 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small molecule AZD4635 inhibitor of A2AR signaling rescues immune cell function including CD103+ dendritic cells enhancing anti-tumor immunity
J Immunother Cancer, 2020
|
|
7EBD
 
 | | Bacterial STING in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ACETATE ION, TIR-like domain-containing protein | | Authors: | Ko, T.-P, Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure and functional implication of bacterial STING.
Nat Commun, 13, 2022
|
|
2NNP
 
 | | Crystal structure analysis of HIV-1 protease mutant I84V with a inhibitor saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, ACETIC ACID, GLYCEROL, ... | | Authors: | Tie, Y, Kovalevsky, A.Y, Boross, P, Wang, Y.F, Ghosh, A.K, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2006-10-24 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir.
Proteins, 67, 2007
|
|
7WDM
 
 | | Fungal immunomodulatory protein FIP-gmi | | Descriptor: | Immunomodulatory protein | | Authors: | Liu, Y, Bastiaan-Net, S, Zhang, Y, Hoppenbrouwers, T, Xie, Y, Wang, Y, Wei, X, Du, G, Zhang, H, Imam, K.M.S.U, Wichers, H.J, Li, Z. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.123 Å) | | Cite: | Linking the thermostability of FIP-nha (Nectria haematococca) to its structural properties
To Be Published
|
|
2NMZ
 
 | | Crystal structure analysis of HIV-1 protease mutant V82A with a inhibitor saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, PROTEASE, SULFATE ION | | Authors: | Tie, Y, Kovalevsky, A.Y, Boross, P, Wang, Y.F, Ghosh, A.K, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2006-10-23 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir.
Proteins, 67, 2007
|
|
2NNK
 
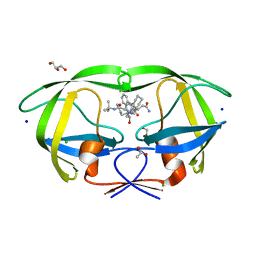 | | Crystal structure analysis of HIV-1 protease mutant I84V with a inhibitor saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Tie, Y, Kovalevsky, A.Y, Boross, P, Wang, Y.F, Ghosh, A.K, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2006-10-24 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir.
Proteins, 67, 2007
|
|
2NMY
 
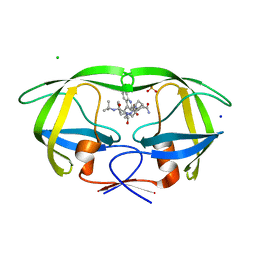 | | Crystal structure analysis of HIV-1 protease mutant V82A with a inhibitor saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, PROTEASE, ... | | Authors: | Tie, Y, Kovalevsky, A.Y, Boross, P, Wang, Y.F, Ghosh, A.K, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2006-10-23 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir.
Proteins, 67, 2007
|
|
3W9K
 
 | | Crystal structure of thermoacidophile-specific protein STK_08120 complexed with myristic acid | | Descriptor: | FATTY ACID-BINDING PROTEIN, MYRISTIC ACID | | Authors: | Miyakawa, T, Sawano, Y, Miyazono, K, Miyauchi, Y, Hatano, K, Tanokura, M. | | Deposit date: | 2013-04-05 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A thermoacidophile-specific protein family, DUF3211, functions as a fatty acid carrier with novel binding mode.
J.Bacteriol., 195, 2013
|
|
4XRE
 
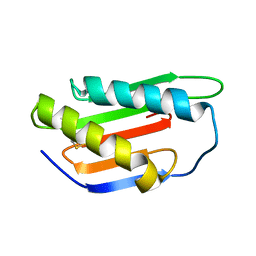 | | Crystal structure of Gnk2 complexed with mannose | | Descriptor: | Antifungal protein ginkbilobin-2, alpha-D-mannopyranose | | Authors: | Miyakawa, T, Hatano, K, Miyauchi, Y, Suwa, Y, Sawano, Y, Tanokura, M. | | Deposit date: | 2015-01-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | A secreted protein with plant-specific cysteine-rich motif functions as a mannose-binding lectin that exhibits antifungal activity.
Plant Physiol., 166, 2014
|
|
7CNO
 
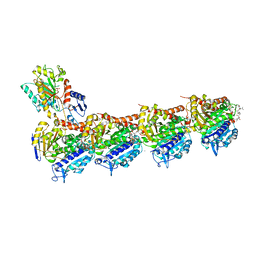 | | Phomopsin A in complex with tubulin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wu, C.Y, Wang, Y.X. | | Deposit date: | 2020-08-02 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The high-resolution X-ray structure of vinca-domain inhibitors of microtubules provides a rational approach for drug design.
Febs Lett., 595, 2021
|
|
7D6H
 
 | |
5ZXH
 
 | | The structure of MT189-tubulin complex | | Descriptor: | 2-(6-fluoro-3-{[(4-methoxyphenyl)methyl]amino}imidazo[1,2-a]pyridin-2-yl)phenol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Li, Z.P, Wang, Y.X, Meng, T, Yang, J.L, Chen, Q. | | Deposit date: | 2018-05-21 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of MT189-Tubulin Complex Provides Insights into Drug Design
Lett.Drug Des.Discovery, 2019
|
|
2CWV
 
 | | Product schiff-base intermediate of copper amine oxidase from arthrobacter globiformis | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Chiu, Y.C, Okajima, T, Murakawa, T, Uchida, M, Taki, M, Hirota, S, Kim, M, Yamaguchi, H, Kawano, Y, Kamiya, N, Kuroda, S, Hayashi, H, Yamamoto, Y, Tanizawa, K. | | Deposit date: | 2005-06-26 | | Release date: | 2006-05-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Kinetic and Structural Studies on the Catalytic Role of the Aspartic Acid Residue Conserved in Copper Amine Oxidase(,)
Biochemistry, 45, 2006
|
|
2CWU
 
 | | Substrate schiff-base intermediate of copper amine oxidase from arthrobacter globiformis | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Chiu, Y.C, Okajima, T, Murakawa, T, Uchida, M, Taki, M, Hirota, S, Kim, M, Yamaguchi, H, Kawano, Y, Kamiya, N, Kuroda, S, Hayashi, H, Yamamoto, Y, Tanizawa, K. | | Deposit date: | 2005-06-26 | | Release date: | 2006-05-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Kinetic and Structural Studies on the Catalytic Role of the Aspartic Acid Residue Conserved in Copper Amine Oxidase(,)
Biochemistry, 45, 2006
|
|
2CWT
 
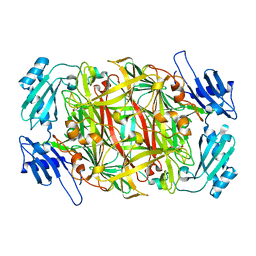 | | Catalytic base deletion in copper amine oxidase from arthrobacter globiformis | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Chiu, Y.C, Okajima, T, Murakawa, T, Uchida, M, Taki, M, Hirota, S, Kim, M, Yamaguchi, H, Kawano, Y, Kamiya, N, Kuroda, S, Hayashi, H, Yamamoto, Y, Tanizawa, K. | | Deposit date: | 2005-06-26 | | Release date: | 2006-05-02 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Kinetic and Structural Studies on the Catalytic Role of the Aspartic Acid Residue Conserved in Copper Amine Oxidase(,)
Biochemistry, 45, 2006
|
|
2ZZD
 
 | | Recombinant thiocyanate hydrolase, air-oxidized form of holo-enzyme | | Descriptor: | COBALT (III) ION, L(+)-TARTARIC ACID, Thiocyanate hydrolase subunit alpha, ... | | Authors: | Arakawa, T, Kawano, Y, Katayama, Y, Yohda, M, Odaka, M. | | Deposit date: | 2009-02-09 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for Catalytic Activation of Thiocyanate Hydrolase Involving Metal-Ligated Cysteine Modification
J.Am.Chem.Soc., 131, 2009
|
|
7C5G
 
 | | Crystal Structure of C150S mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with PO4 at 1.98 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
2H7F
 
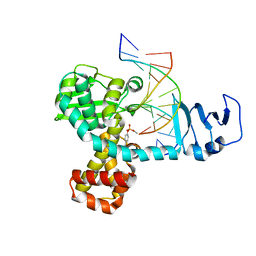 | | Structure of variola topoisomerase covalently bound to DNA | | Descriptor: | 5'-D(*TP*AP*AP*TP*AP*AP*GP*GP*GP*CP*GP*AP*CP*A)-3', 5'-D(*TP*TP*GP*TP*CP*GP*CP*CP*CP*TP*T)-3', DNA topoisomerase 1 | | Authors: | Perry, K, Hwang, Y, Bushman, F.D, Van Duyne, G.D. | | Deposit date: | 2006-06-02 | | Release date: | 2006-08-15 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for specificity in the poxvirus topoisomerase.
Mol.Cell, 23, 2006
|
|
7E35
 
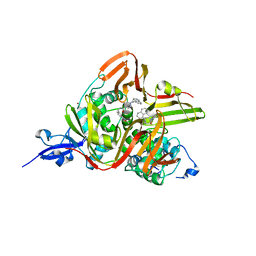 | | Crystal structure of the SARS-CoV-2 papain-like protease (PLPro) C112S mutant bound to compound S43 | | Descriptor: | N-[(3-acetamidophenyl)methyl]-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide, Non-structural protein 3, ZINC ION | | Authors: | Liu, J, Wang, Y, Xu, X, Pan, L. | | Deposit date: | 2021-02-08 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development of potent and selective inhibitors targeting the papain-like protease of SARS-CoV-2.
Cell Chem Biol, 28, 2021
|
|
2H7G
 
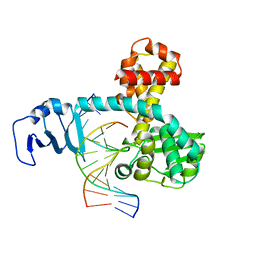 | | Structure of variola topoisomerase non-covalently bound to DNA | | Descriptor: | 5'-D(*TP*AP*AP*TP*AP*AP*GP*GP*GP*CP*GP*AP*CP*A)-3', 5'-D(*TP*TP*GP*TP*CP*GP*CP*CP*CP*TP*TP*A)-3', DNA topoisomerase 1 | | Authors: | Perry, K, Hwang, Y, Bushman, F.D, Van Duyne, G.D. | | Deposit date: | 2006-06-02 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for specificity in the poxvirus topoisomerase.
Mol.Cell, 23, 2006
|
|
1X25
 
 | | Crystal Structure of a Member of YjgF Family from Sulfolobus Tokodaii (ST0811) | | Descriptor: | Hypothetical UPF0076 protein ST0811 | | Authors: | Miyakawa, T, Lee, W.C, Hatano, K, Kato, Y, Sawano, Y, Miyazono, K, Nagata, K, Tanokura, M. | | Deposit date: | 2005-04-20 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the YjgF/YER057c/UK114 family protein from the hyperthermophilic archaeon Sulfolobus tokodaii strain 7
Proteins, 62, 2006
|
|
7C5O
 
 | | Crystal Structure of H177A mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with NAD at 1.98 Angstrom resolution. | | Descriptor: | CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5K
 
 | | Crystal Structure of C150S mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with G3P at 2.69 Angstrom resolution | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, GLYCERALDEHYDE-3-PHOSPHATE, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5L
 
 | | Crystal Structure of C150S mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli at 2.1 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
