8P4O
 
 | | CryoEM structure of a GroEL7-GroES7 cage with encapsulated ordered substrate MetK in the presence of ADP-BeFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Wagner, J, Beck, F, Bracher, A, Caravajal, A.I, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-05-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
7ELG
 
 | | LC3B modificated with a covalent probe | | Descriptor: | 2-methylidene-5-thiophen-2-yl-cyclohexane-1,3-dione, Microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Fan, S, Wan, W. | | Deposit date: | 2021-04-10 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Inhibition of Autophagy by a Small Molecule through Covalent Modification of the LC3 Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6W3E
 
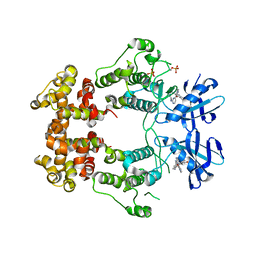 | | Structure of phosphorylated IRE1 in complex with G-0701 | | Descriptor: | Serine/threonine-protein kinase/endoribonuclease IRE1, methyl ~{N}-[6-methyl-5-[3-[2-[[(3~{S})-piperidin-3-yl]amino]pyrimidin-4-yl]pyridin-2-yl]oxy-naphthalen-1-yl]carbamate | | Authors: | Ferri, E, Wang, W, Joachim, R, Mortara, K. | | Deposit date: | 2020-03-09 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.737 Å) | | Cite: | Activation of the IRE1 RNase through remodeling of the kinase front pocket by ATP-competitive ligands.
Nat Commun, 11, 2020
|
|
6W39
 
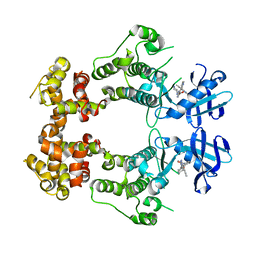 | | Structure of unphosphorylated IRE1 in complex with G-1749 | | Descriptor: | Serine/threonine-protein kinase/endoribonuclease IRE1, ethyl ~{N}-[6-methyl-5-[3-[2-[[(3~{S})-piperidin-3-yl]amino]pyrimidin-4-yl]pyridin-2-yl]oxy-naphthalen-1-yl]carbamate | | Authors: | Ferri, E, Wang, W, Joachim, R, Mortara, K. | | Deposit date: | 2020-03-09 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.736 Å) | | Cite: | Activation of the IRE1 RNase through remodeling of the kinase front pocket by ATP-competitive ligands.
Nat Commun, 11, 2020
|
|
6W3B
 
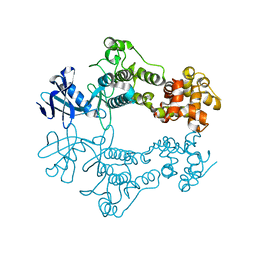 | | Structure of apo unphosphorylated IRE1 | | Descriptor: | Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Wallweber, H, Mortara, K, Ferri, E, Rudolph, J, Wang, W. | | Deposit date: | 2020-03-09 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Activation of the IRE1 RNase through remodeling of the kinase front pocket by ATP-competitive ligands.
Nat Commun, 11, 2020
|
|
4ZY4
 
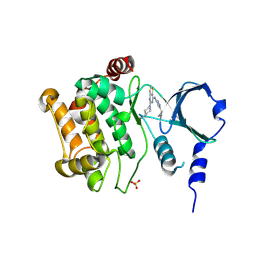 | |
4ZY6
 
 | |
4ZY5
 
 | |
4G5S
 
 | | Structure of LGN GL3/Galphai3 complex | | Descriptor: | CITRIC ACID, G-protein-signaling modulator 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jia, M, Li, J, Zhu, J, Wen, W, Zhang, M, Wang, W. | | Deposit date: | 2012-07-18 | | Release date: | 2012-09-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Crystal Structures of the scaffolding protein LGN reveal the general mechanism by which GoLoco binding motifs inhibit the release of GDP from Galphai subunits in G-coupled heterotrimeric proteins
To be Published
|
|
3UV6
 
 | | Ec_IspH in complex with 4-hydroxybutyl diphosphate (1301) | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, 4-hydroxybutyl trihydrogen diphosphate, IRON/SULFUR CLUSTER | | Authors: | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Schulz, C, Oldfield, E, Groll, M. | | Deposit date: | 2011-11-29 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
3UWM
 
 | | Ec_IspH in complex with 4-oxobutyl diphosphate (1302) | | Descriptor: | (3E)-4-hydroxybut-3-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, 4-oxobutyl trihydrogen diphosphate, ... | | Authors: | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Schulz, C, Oldfield, E, Groll, M. | | Deposit date: | 2011-12-02 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
4G2V
 
 | | Structure complex of LGN binding with FRMPD1 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, G-protein-signaling modulator 2, ... | | Authors: | Shang, Y, Pan, Z, Wen, W, Wang, W, Zhang, M. | | Deposit date: | 2012-07-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biochemical characterization of the interaction between LGN and Frmpd1
J.Mol.Biol., 425, 2013
|
|
4G5R
 
 | | Structure of LGN GL4/Galphai3 complex | | Descriptor: | CITRIC ACID, G-protein-signaling modulator 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jia, M, Li, J, Zhu, J, Wen, W, Zhang, M, Wang, W. | | Deposit date: | 2012-07-18 | | Release date: | 2012-09-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.481 Å) | | Cite: | Crystal Structures of the scaffolding protein LGN reveal the general mechanism by which GoLoco binding motifs inhibit the release of GDP from Galphai subunits in G-coupled heterotrimeric proteins
To be Published
|
|
4G5O
 
 | | Structure of LGN GL4/Galphai3(Q147L) complex | | Descriptor: | CITRIC ACID, G-protein-signaling modulator 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jia, M, Li, J, Zhu, J, Wen, W, Zhang, M, Wang, W. | | Deposit date: | 2012-07-18 | | Release date: | 2012-09-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of the scaffolding protein LGN reveal the general mechanism by which GoLoco binding motifs inhibit the release of GDP from Galphai subunits in G-coupled heterotrimeric proteins
To be Published
|
|
4G5Q
 
 | | Structure of LGN GL4/Galphai1 complex | | Descriptor: | CITRIC ACID, G-protein-signaling modulator 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jia, M, Li, J, Zhu, J, Wen, W, Zhang, M, Wang, W. | | Deposit date: | 2012-07-18 | | Release date: | 2012-09-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of the scaffolding protein LGN reveal the general mechanism by which GoLoco binding motifs inhibit the release of GDP from Galphai subunits in G-coupled heterotrimeric proteins
To be Published
|
|
1N94
 
 | | Aryl Tetrahydropyridine Inhbitors of Farnesyltransferase: Glycine, Phenylalanine and Histidine Derivates | | Descriptor: | 2-{(5-{[BUTYL-(2-CYCLOHEXYL-ETHYL)-AMINO]-METHYL}-2'-METHYL-BIPHENYL-2-CARBONYL)-AMINO]-4-METHYLSULFANYL-BUTYRIC ACID, ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID, Protein farnesyltransferase alpha subunit, ... | | Authors: | Gwaltney II, S.L, O'Connor, S.J, Nelson, L.T, Sullivan, G.M, Imade, H, Wang, W, Hasvold, L, Li, Q, Cohen, J, Gu, W.Z. | | Deposit date: | 2002-11-22 | | Release date: | 2003-01-07 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Aryl tetrahydropyridine inhibitors of farnesyltransferase: glycine, phenylalanine and histidine derivatives.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1N9A
 
 | | Farnesyltransferase complex with tetrahydropyridine inhibitors | | Descriptor: | 1-{2-[3-(4-CYANO-BENZYL)-3H-IMIDAZOL-4-YL]-ACETYL}-5-NAPHTHALEN-1-YL-1,2,3,6-TETRAHYDRO-PYRIDINE-4-CARBONITRILE, ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID, Protein farnesyltransferase alpha subunit, ... | | Authors: | Gwaltney II, S.L, O'Conner, S.J, Nelson, L.T, Sullivan, G.M, Imade, H, Wang, W, Hasvold, L, Li, Q, Cohen, J, Gu, W.Z. | | Deposit date: | 2002-11-22 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Aryl tetrahydropyridine inhibitors of farnesyltransferase: bioavailable analogues with improved cellular potency.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
5W96
 
 | | Solution structure of phage derived peptide inhibitor of frizzled 7 receptor | | Descriptor: | Fz7 binding peptide | | Authors: | Nile, A.H, de Sousa e Melo, F, Mukund, S, Piskol, R, Hansen, S, Zhou, L, Zhang, Y, Fu, Y, Gogol, E.B, Komuves, L.G, Modrusan, Z, Angers, S, Franke, Y, Koth, C, Fairbrother, W.J, Wang, W, de Sauvage, F.J, Hannoush, R.N. | | Deposit date: | 2017-06-22 | | Release date: | 2018-04-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A selective peptide inhibitor of Frizzled 7 receptors disrupts intestinal stem cells.
Nat. Chem. Biol., 14, 2018
|
|
6CQD
 
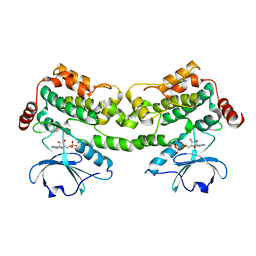 | | Crystal structure of HPK1 in complex with ATP analogue (AMPPNP) | | Descriptor: | MAGNESIUM ION, Mitogen-activated protein kinase kinase kinase kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wu, P, Lehoux, I, Franke, Y, Mortara, K, Wang, W. | | Deposit date: | 2018-03-14 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Hematopoietic Progenitor Kinase-1 Structure in a Domain-Swapped Dimer.
Structure, 27, 2019
|
|
6CQE
 
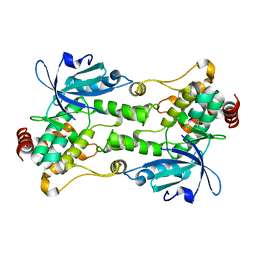 | | Crystal structure of HPK1 kinase domain S171A mutant | | Descriptor: | Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Wu, P, Lehoux, I, Mortara, K, Franke, Y, Wang, W. | | Deposit date: | 2018-03-15 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.886 Å) | | Cite: | Hematopoietic Progenitor Kinase-1 Structure in a Domain-Swapped Dimer.
Structure, 27, 2019
|
|
4FXX
 
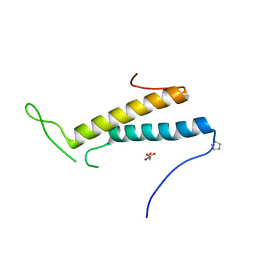 | | Structure of SF1 coiled-coil domain | | Descriptor: | IMIDAZOLE, MALONATE ION, Splicing factor 1 | | Authors: | Gupta, A, Bauer, W.J, Wang, W, Kielkopf, C.L. | | Deposit date: | 2012-07-03 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4801 Å) | | Cite: | Structure of Phosphorylated SF1 Bound to U2AF(65) in an Essential Splicing Factor Complex.
Structure, 21, 2013
|
|
4G9Z
 
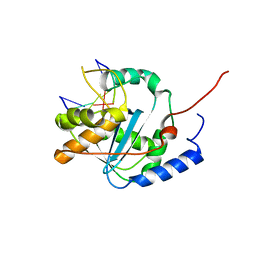 | | Lassa nucleoprotein with dsRNA reveals novel mechanism for immune suppression | | Descriptor: | Nucleoprotein, RNA (5'-R(P*GP*CP*CP*C)-3'), RNA (5'-R(P*GP*GP*GP*C)-3'), ... | | Authors: | Jiang, X, Huang, Q, Wang, W, Dong, H, Ly, H, Liang, Y, Dong, C, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2012-07-24 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of Arenaviral Nucleoproteins with Triphosphate dsRNA Reveal a Unique Mechanism of Immune Suppression.
J.Biol.Chem., 288, 2013
|
|
8I79
 
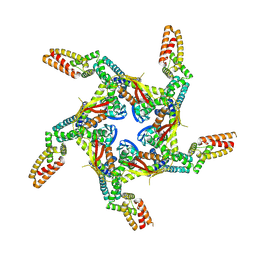 | | Cryo-EM structure of KCTD7 in complex with Cullin3 | | Descriptor: | BTB/POZ domain-containing protein KCTD7, Cullin-3 | | Authors: | Jiang, W, Wang, W, Zheng, S. | | Deposit date: | 2023-01-31 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for the ubiquitination of G protein beta gamma subunits by KCTD5/Cullin3 E3 ligase.
Sci Adv, 9, 2023
|
|
6W3A
 
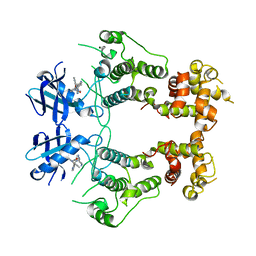 | | Structure of unphosphorylated IRE1 in complex with G-7658 | | Descriptor: | Serine/threonine-protein kinase/endoribonuclease IRE1, ~{N}-[6-methyl-5-[3-[2-[[(3~{S})-piperidin-3-yl]amino]pyrimidin-4-yl]pyridin-2-yl]oxy-naphthalen-1-yl]but-2-ynamide | | Authors: | Ferri, E, Wang, W, Joachim, R, Mortara, K. | | Deposit date: | 2020-03-09 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Activation of the IRE1 RNase through remodeling of the kinase front pocket by ATP-competitive ligands.
Nat Commun, 11, 2020
|
|
6W3C
 
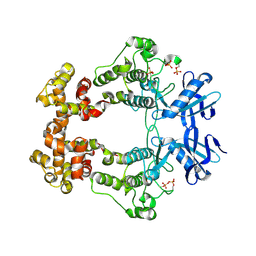 | | Structure of phosphorylated apo IRE1 | | Descriptor: | Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Wallweber, H, Mortara, K, Ferri, E, Wang, W, Rudolph, J. | | Deposit date: | 2020-03-09 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Activation of the IRE1 RNase through remodeling of the kinase front pocket by ATP-competitive ligands.
Nat Commun, 11, 2020
|
|
