6LSF
 
 | | Crystal structure of the enterovirus 71 polymerase elongation complex (C2S6RA/C2S6RB form) | | Descriptor: | Genome polyprotein, RNA (35-MER), RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*GP*AP*GP*AP*GP*AP*CP*C)-3'), ... | | Authors: | Wang, M, Shu, B, Jing, X, Gong, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Stringent control of the RNA-dependent RNA polymerase translocation revealed by multiple intermediate structures.
Nat Commun, 11, 2020
|
|
6LSE
 
 | | Crystal structure of the enterovirus 71 polymerase elongation complex (C3S6A/C3S6B form) | | Descriptor: | Genome polyprotein, PYROPHOSPHATE 2-, RNA (35-MER), ... | | Authors: | Wang, M, Shu, B, Jing, X, Gong, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Stringent control of the RNA-dependent RNA polymerase translocation revealed by multiple intermediate structures.
Nat Commun, 11, 2020
|
|
5VZ1
 
 | | Crystal structure of the Apo Antibody fragment (Fab) raised against C-terminal domain of Ebola nucleoprotein (EBOV, TAFV, BDBV strains) | | Descriptor: | Apo Antibody Fab Heavy Chain, Apo Antibody Fab Light Chain, ZINC ION | | Authors: | Radwanska, M.J, Derewenda, U, Kossiakoff, A, Derewenda, Z.S. | | Deposit date: | 2017-05-26 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | Crystal structure of the Apo Antibody fragment (Fab) raised against C-terminal domain of Ebola nucleoprotein (EBOV, TAFV, BDBV strains)
To Be Published
|
|
3IAY
 
 | | Ternary complex of DNA polymerase delta | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*AP*TP*CP*CP*TP*CP*CP*CP*CP*TP*AP*(DOC))-3', 5'-D(*TP*AP*AP*GP*GP*TP*AP*GP*GP*GP*GP*AP*GP*GP*AP*T)-3', ... | | Authors: | Swan, M.K, Aggarwal, A.K. | | Deposit date: | 2009-07-15 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta
Nat.Struct.Mol.Biol., 16, 2009
|
|
3GQC
 
 | | Structure of human Rev1-DNA-dNTP ternary complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*AP*TP*CP*CP*TP*CP*CP*CP*CP*TP*AP*(DOC))-3', 5'-D(*TP*AP*AP*GP*GP*TP*AP*GP*GP*GP*GP*AP*GP*GP*AP*T)-3', ... | | Authors: | Swan, M.K, Aggarwal, A.K. | | Deposit date: | 2009-03-24 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the human Rev1-DNA-dNTP ternary complex.
J.Mol.Biol., 390, 2009
|
|
1X9I
 
 | | Crystal structure of Crystal structure of phosphoglucose/phosphomannose phosphoglucose/phosphomannoseisomerase from Pyrobaculum aerophilum in complex with glucose 6-phosphate | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLYCEROL, glucose-6-phosphate isomerase | | Authors: | Swan, M.K, Hansen, T, Schoenheit, P, Davies, C. | | Deposit date: | 2004-08-21 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structural basis for phosphomannose isomerase activity in phosphoglucose isomerase from Pyrobaculum aerophilum: a subtle difference between distantly related enzymes.
Biochemistry, 43, 2004
|
|
1QY4
 
 | | Crystal structure of phosphoglucose isomerase from Pyrococcus furiosus in complex with gluconate 6-phosphate | | Descriptor: | 6-PHOSPHOGLUCONIC ACID, Glucose-6-phosphate isomerase, NICKEL (II) ION | | Authors: | Swan, M.K, Solomons, J.T.G, Beeson, C.C, Hansen, T, Schonheit, P, Davies, C. | | Deposit date: | 2003-09-09 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evidence for a hydride transfer mechanism of catalysis in phosphoglucose isomerase from Pyrococcus furiosus
J.Biol.Chem., 278, 2003
|
|
1QXJ
 
 | | Crystal structure of native phosphoglucose isomerase from Pyrococcus furiosus | | Descriptor: | Glucose-6-phosphate isomerase, NICKEL (II) ION | | Authors: | Swan, M.K, Solomons, J.T.G, Beeson, C.C, Hansen, T, Schonheit, P, Davies, C. | | Deposit date: | 2003-09-07 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evidence for a hydride transfer mechanism of catalysis in phosphoglucose isomerase from Pyrococcus furiosus
J.Biol.Chem., 278, 2003
|
|
1QXR
 
 | | Crystal structure of phosphoglucose isomerase from Pyrococcus furiosus in complex with 5-phosphoarabinonate | | Descriptor: | 5-PHOSPHOARABINONIC ACID, Glucose-6-phosphate isomerase, NICKEL (II) ION | | Authors: | Swan, M.K, Solomons, J.T.G, Beeson, C.C, Hansen, P, Schonheit, P, Davies, C. | | Deposit date: | 2003-09-08 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural evidence for a hydride transfer mechanism of catalysis in phosphoglucose isomerase from Pyrococcus furiosus
J.Biol.Chem., 278, 2003
|
|
7WC2
 
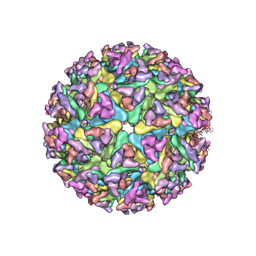 | | Cryo-EM structure of alphavirus, Getah virus | | Descriptor: | Spike glycoprotein E1, Spike glycoprotein E2, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, M, Sun, Z.Z, Wang, J.F. | | Deposit date: | 2021-12-18 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Implications for the pathogenicity and antigenicity of alpha viruses revealed by a 3.5 angstrom Cryo-EM structure of Getah virus
To Be Published
|
|
6B6Z
 
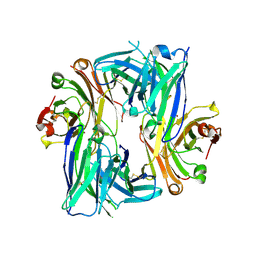 | | Crystal structure of the Apo Antibody fragment (Fab) raised against C-terminal domain of Ebola nucleoprotein (EBOV, TAFV, BDBV strains) | | Descriptor: | Apo Fab Heavy Chain, Apo Fab Light Chain, ZINC ION | | Authors: | Radwanska, M.J, Derewenda, U, Kossiakoff, A.A, Derewenda, Z.S. | | Deposit date: | 2017-10-03 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | Crystal structure of the Apo Antibody fragment (Fab) raised against C-terminal domain of Ebola nucleoprotein (EBOV, TAFV, BDBV strains)
To Be Published
|
|
7WCO
 
 | | Cryo-EM structure of alphavirus, Getah virus | | Descriptor: | Capsid protein, Spike glycoprotein E1, Spike glycoprotein E2 | | Authors: | Wang, M, Sun, Z.Z, Wang, J.F. | | Deposit date: | 2021-12-20 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Implications for the pathogenicity and antigenicity of alpha viruses revealed by a 3.5 angstrom Cryo-EM structure of Getah virus
To Be Published
|
|
2NRA
 
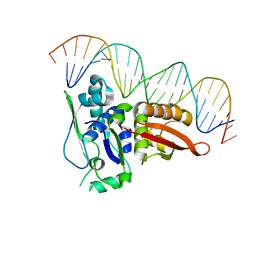 | | Crystal structure of Pi initiator protein in complex with iteron DNA | | Descriptor: | 5'-D(*GP*AP*AP*CP*AP*TP*GP*AP*GP*AP*GP*CP*TP*TP*AP*GP*TP*AP*CP*GP*TP*CP*T)-3', 5'-D(*GP*AP*CP*GP*TP*AP*CP*TP*AP*AP*GP*CP*TP*CP*TP*CP*AP*TP*GP*TP*TP*CP*T)-3', PI protein | | Authors: | Swan, M.K, Bastia, D, Davies, C. | | Deposit date: | 2006-11-01 | | Release date: | 2006-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of pi initiator protein-iteron complex of plasmid R6K: implications for initiation of plasmid DNA replication.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
7W9S
 
 | |
1T8P
 
 | | Crystal structure of Human erythrocyte 2,3-bisphosphoglycerate mutase | | Descriptor: | Bisphosphoglycerate mutase | | Authors: | Wang, Y, Wei, Z, Bian, Q, Cheng, Z, Wan, M, Liu, L, Gong, W. | | Deposit date: | 2004-05-13 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human bisphosphoglycerate mutase
J.Biol.Chem., 279, 2004
|
|
4WBB
 
 | | Single Turnover Autophosphorylation Cycle of the PKA RIIb Holoenzyme | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Zhang, P, Knape, M.J, Ahuja, L.G, Keshwani, M.M, King, C.C, Sastri, M, Herberg, F.W, Taylor, S.S. | | Deposit date: | 2014-09-02 | | Release date: | 2015-05-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Single Turnover Autophosphorylation Cycle of the PKA RII beta Holoenzyme.
Plos Biol., 13, 2015
|
|
4WBQ
 
 | | Crystal structure of the exonuclease domain of QIP (QDE-2 interacting protein) solved by native-SAD phasing. | | Descriptor: | CALCIUM ION, QDE-2-interacting protein | | Authors: | Boland, A, Weinert, T, Weichenrieder, O, Wang, M. | | Deposit date: | 2014-09-03 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4WBX
 
 | | Conserved hypothetical protein PF1771 from Pyrococcus furiosus solved by sulfur SAD using Swiss Light Source data | | Descriptor: | 2-keto acid:ferredoxin oxidoreductase subunit alpha | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-09-04 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
5V54
 
 | | Crystal structure of 5-HT1B receptor in complex with methiothepin | | Descriptor: | 1-methyl-4-[(5~{S})-3-methylsulfanyl-5,6-dihydrobenzo[b][1]benzothiepin-5-yl]piperazine, 5-hydroxytryptamine receptor 1B,OB-1 fused 5-HT1b receptor,5-hydroxytryptamine receptor 1B | | Authors: | Yin, W.C, Zhou, X.E, Yang, D, de Waal, P, Wang, M.T, Dai, A, Cai, X, Huang, C.Y, Liu, P, Yin, Y, Liu, B, Caffrey, M, Melcher, K, Xu, Y, Wang, M.W, Xu, H.E, Jiang, Y. | | Deposit date: | 2017-03-13 | | Release date: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | A common antagonistic mechanism for class A GPCRs revealed by the structure of the human 5-HT1B serotonin receptor bound to an antagonist
Cell Discov, 2018
|
|
1T7B
 
 | | Crystal structure of mutant Lys8Gln of scorpion alpha-like neurotoxin BmK M1 from Buthus martensii Karsch | | Descriptor: | Alpha-like neurotoxin BmK-I | | Authors: | Xiang, Y, Guan, R.J, He, X.L, Wang, C.G, Wang, M, Zhang, Y, Sundberg, E.J, Wang, D.C. | | Deposit date: | 2004-05-09 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Mechanism Governing Cis and Trans Isomeric States and an Intramolecular Switch for Cis/Trans Isomerization of a Non-proline Peptide Bond Observed in Crystal Structures of Scorpion Toxins
J.Mol.Biol., 341, 2004
|
|
1T7A
 
 | | Crystal structure of mutant Lys8Asp of scorpion alpha-like neurotoxin BmK M1 from Buthus martensii Karsch | | Descriptor: | Alpha-like neurotoxin BmK-I | | Authors: | Xiang, Y, Guan, R.J, He, X.L, Wang, C.G, Wang, M, Zhang, Y, Sundberg, E.J, Wang, D.C. | | Deposit date: | 2004-05-08 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Mechanism Governing Cis and Trans Isomeric States and an Intramolecular Switch for Cis/Trans Isomerization of a Non-proline Peptide Bond Observed in Crystal Structures of Scorpion Toxins
J.Mol.Biol., 341, 2004
|
|
1T7E
 
 | | Crystal structure of mutant Pro9Ser of scorpion alpha-like neurotoxin BmK M1 from Buthus martensii Karsch | | Descriptor: | Alpha-like neurotoxin BmK-I, PHOSPHATE ION | | Authors: | Xiang, Y, Guan, R.J, He, X.L, Wang, C.G, Wang, M, Zhang, Y, Sundberg, E.J, Wang, D.C. | | Deposit date: | 2004-05-09 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Mechanism Governing Cis and Trans Isomeric States and an Intramolecular Switch for Cis/Trans Isomerization of a Non-proline Peptide Bond Observed in Crystal Structures of Scorpion Toxins
J.Mol.Biol., 341, 2004
|
|
7AT6
 
 | | Structure of thaumatin collected by femtosecond serial crystallography on a COC membrane | | Descriptor: | L(+)-TARTARIC ACID, R-1,2-PROPANEDIOL, SODIUM ION, ... | | Authors: | Martiel, I, Marsh, M, Vera, L, Huang, C.Y, Olieric, V, Leonarski, P, Nass, K, Padeste, C, Karpik, A, Wang, M, Pedrini, B. | | Deposit date: | 2020-10-29 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Commissioning results from the SwissMX instrument for fixed target macromolecular crystallography at SwissFEL
To Be Published
|
|
7AI8
 
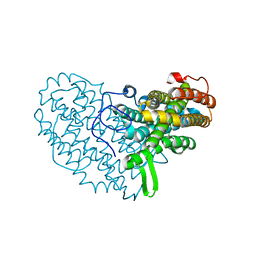 | | Structure of Ribonucleotide reductase R2 from Escherichia coli collected by still serial crystallography on a COC membrane at a synchrotron source | | Descriptor: | FE (III) ION, Ribonucleoside-diphosphate reductase 1 subunit beta | | Authors: | Aurelius, O, John, J, Martiel, I, Padeste, C, Karpik, A, Huang, C.Y, Hogbom, M, Wang, M, Marsh, M. | | Deposit date: | 2020-09-26 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Versatile microporous polymer-based supports for serial macromolecular crystallography.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7AI9
 
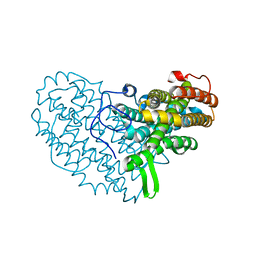 | | Structure of Ribonucleotide reductase R2 from Escherichia coli collected by rotation serial crystallography on a COC membrane at a synchrotron source | | Descriptor: | FE (III) ION, Ribonucleoside-diphosphate reductase 1 subunit beta | | Authors: | Aurelius, O, John, J, Martiel, I, Padeste, C, Karpik, A, Huang, C.Y, Hogbom, M, Wang, M, Marsh, M. | | Deposit date: | 2020-09-26 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Versatile microporous polymer-based supports for serial macromolecular crystallography.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
