4EK7
 
 | | High speed X-ray analysis of plant enzymes at room temperature | | Descriptor: | CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase, beta-D-glucopyranose | | Authors: | Xia, L, Rajendran, C, Ruppert, M, Panjikar, S, Wang, M, Stoeckigt, J. | | Deposit date: | 2012-04-09 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High speed X-ray analysis of plant enzymes at room temperature.
Phytochemistry, 2012
|
|
4FYP
 
 | | Crystal Structure of Plant Vegetative Storage Protein | | Descriptor: | MAGNESIUM ION, Vegetative storage protein 1 | | Authors: | Chen, Y, Wei, J, Wang, M, Gong, W, Zhang, M. | | Deposit date: | 2012-07-05 | | Release date: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of Arabidopsis VSP1 reveals the plant class C-like phosphatase structure of the DDDD superfamily of phosphohydrolases
Plos One, 7, 2012
|
|
4PW6
 
 | | structure of UHRF2-SRA in complex with a 5hmC-containing DNA, complex II | | Descriptor: | 5hmC-containing DNA1, 5hmC-containing DNA2, E3 ubiquitin-protein ligase UHRF2 | | Authors: | Zhou, T, Xiong, J, Wang, M, Yang, N, Wong, J, Zhu, B, Xu, R.M. | | Deposit date: | 2014-03-18 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.789 Å) | | Cite: | Structural Basis for Hydroxymethylcytosine Recognition by the SRA Domain of UHRF2.
Mol.Cell, 54, 2014
|
|
8IOW
 
 | |
8JIP
 
 | | Cryo-EM structure of the GLP-1R/GCGR dual agonist MEDI0382-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, L, Zhou, Q.T, Dai, A.T, Zhao, F.H, Chang, R.L, Ying, T.L, Wu, B.L, Yang, D.H, Wang, M.W, Cong, Z.T. | | Deposit date: | 2023-05-27 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural analysis of the dual agonism at GLP-1R and GCGR.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3U57
 
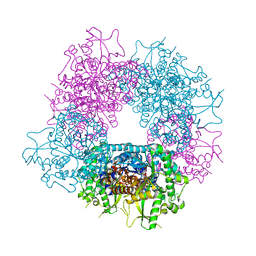 | | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity | | Descriptor: | (2beta,7beta,16S,17R,19E,21beta)-21-(beta-D-glucopyranosyloxy)-2,7-dihydro-7,17-cyclosarpagan-17-yl acetate, CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase | | Authors: | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Lin, H, Rajendran, C, Barleben, L, Stoeckigt, J. | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structures of alkaloid biosynthetic glucosidases decode substrate specificity.
Acs Chem.Biol., 7, 2012
|
|
8JIR
 
 | | Cryo-EM structure of the GLP-1R/GCGR dual agonist SAR425899-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, L, Zhou, Q.T, Dai, A.T, Zhao, F.H, Chang, R.L, Ying, T.L, Wu, B.L, Yang, D.H, Wang, M.W, Cong, Z.T. | | Deposit date: | 2023-05-27 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural analysis of the dual agonism at GLP-1R and GCGR.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3U5Y
 
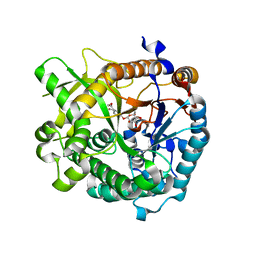 | | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity | | Descriptor: | CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase, Secologanin | | Authors: | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Lin, H, Rajendran, C, Barleben, L, Stoeckigt, J. | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of alkaloid biosynthetic glucosidases decode substrate specificity.
Acs Chem.Biol., 7, 2012
|
|
8JIT
 
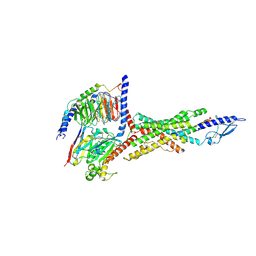 | | Cryo-EM structure of the GLP-1R/GCGR dual agonist MEDI0382-bound human GCGR-Gs complex | | Descriptor: | Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, L, Zhou, Q.T, Dai, A.T, Zhao, F.H, Chang, R.L, Ying, T.L, Wu, B.L, Yang, D.H, Wang, M.W, Cong, Z.T. | | Deposit date: | 2023-05-27 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural analysis of the dual agonism at GLP-1R and GCGR.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JIQ
 
 | | Cryo-EM structure of the GLP-1R/GCGR dual agonist Peptide 15-bound human GCGR-Gs complex | | Descriptor: | Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, L, Zhou, Q.T, Dai, A.T, Zhao, F.H, Chang, R.L, Ying, T.L, Wu, B.L, Yang, D.H, Wang, M.W, Cong, Z.T. | | Deposit date: | 2023-05-27 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural analysis of the dual agonism at GLP-1R and GCGR.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JIU
 
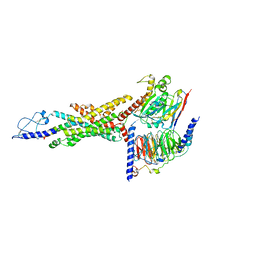 | | Cryo-EM structure of the GLP-1R/GCGR dual agonist SAR425899-bound human GCGR-Gs complex | | Descriptor: | Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, L, Zhou, Q.T, Dai, A.T, Zhao, F.H, Chang, R.L, Ying, T.L, Wu, B.L, Yang, D.H, Wang, M.W, Cong, Z.T. | | Deposit date: | 2023-05-27 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural analysis of the dual agonism at GLP-1R and GCGR.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7C3M
 
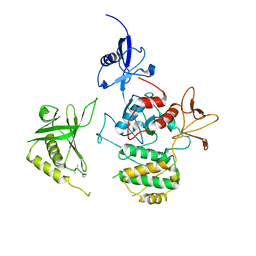 | | Structure of FERM protein | | Descriptor: | Fermitin family homolog 3,Fermitin family homolog 3,Fermitin family homolog 3 | | Authors: | Bu, W, Loh, Z.Y, Jin, S, Basu, S, Ero, R, Park, J.E, Yan, X, Wang, M, Sze, S.K, Tan, S.M, Gao, Y.G. | | Deposit date: | 2020-05-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis of human full-length kindlin-3 homotrimer in an auto-inhibited state.
Plos Biol., 18, 2020
|
|
8JIS
 
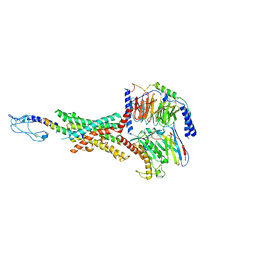 | | Cryo-EM structure of the GLP-1R/GCGR dual agonist peptide15-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, L, Zhou, Q.T, Dai, A.T, Zhao, F.H, Chang, R.L, Ying, T.L, Wu, B.L, Yang, D.H, Wang, M.W, Cong, Z.T. | | Deposit date: | 2023-05-27 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structural analysis of the dual agonism at GLP-1R and GCGR.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6LI6
 
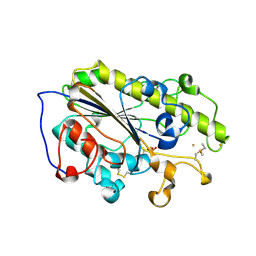 | | Crystal structure of MCR-1-S treated by Au(PEt3)Cl | | Descriptor: | GOLD ION, Probable phosphatidylethanolamine transferase Mcr-1, TRIETHYLPHOSPHANE | | Authors: | Zhang, Q, Wang, M, Sun, H. | | Deposit date: | 2019-12-10 | | Release date: | 2020-09-16 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin.
Nat Commun, 11, 2020
|
|
4EQ3
 
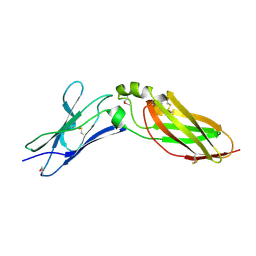 | | Crystal Structure Analysis of Selenomethionine (Se-Met) Substituted Chicken Interferon Gamma Receptor Alpha Chain | | Descriptor: | Interferon gamma receptor 1 | | Authors: | Ping, Z, Qi, J, Lu, G, Shi, Y, Wang, X, Gao, G.F, Wang, M. | | Deposit date: | 2012-04-18 | | Release date: | 2013-04-24 | | Last modified: | 2014-05-21 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of the interferon gamma receptor alpha chain from chicken reveals an undetected extra helix compared with the human counterparts.
J.Interferon Cytokine Res., 34, 2014
|
|
6LHE
 
 | | Crystal Structure of Gold-bound NDM-1 | | Descriptor: | GOLD ION, Metallo-beta-lactamase type 2, SULFATE ION | | Authors: | Wang, H, Sun, H, Wang, M. | | Deposit date: | 2019-12-07 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.206 Å) | | Cite: | Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin.
Nat Commun, 11, 2020
|
|
3JUI
 
 | | Crystal Structure of the C-terminal Domain of Human Translation Initiation Factor eIF2B epsilon Subunit | | Descriptor: | GLYCEROL, Translation initiation factor eIF-2B subunit epsilon | | Authors: | Wei, J, Xu, H, Zhang, C, Wang, M, Gao, F, Gong, W. | | Deposit date: | 2009-09-15 | | Release date: | 2009-10-06 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the C-terminal Domain of Human Translation Initiation Factor eIF2B epsilon Subunit
To be published
|
|
3V0T
 
 | | Crystal Structure of Perakine Reductase, Founder Member of a Novel AKR Subfamily with Unique Conformational Changes during NADPH Binding | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Perakine Reductase | | Authors: | Sun, L, Chen, Y, Rajendran, C, Panjikar, S, Mueller, U, Wang, M, Rosenthal, C, Mindnich, R, Penning, T.M, Stoeckigt, J. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.333 Å) | | Cite: | Crystal structure of perakine reductase, founding member of a novel aldo-keto reductase (AKR) subfamily that undergoes unique conformational changes during NADPH binding.
J.Biol.Chem., 287, 2012
|
|
7DUR
 
 | | Cryo-EM structure of the compound 2-bound human GLP-1 receptor-Gs complex | | Descriptor: | CHOLESTEROL, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z, Chen, L, Ma, H, Zhou, Q, Zou, X, Ye, C, Dai, A, Liu, Q, Huang, W, Sun, X, Wang, X, Xu, P, Zhao, L, Xia, T, Zhong, W, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2021-01-11 | | Release date: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor.
Nat Commun, 12, 2021
|
|
6LI4
 
 | | Crystal structure of MCR-1-S | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION | | Authors: | Zhang, Q, Wang, M, Sun, H. | | Deposit date: | 2019-12-10 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin.
Nat Commun, 11, 2020
|
|
6LI5
 
 | | Crystal structure of apo-MCR-1-S | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1 | | Authors: | Zhang, Q, Wang, M, Sun, H. | | Deposit date: | 2019-12-10 | | Release date: | 2020-09-16 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin.
Nat Commun, 11, 2020
|
|
4GIQ
 
 | | Crystal Structure of mouse RANK bound to RANKL | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, SODIUM ION, ... | | Authors: | Nelson, C.A, Wang, M.W.-H, Fremont, D.H. | | Deposit date: | 2012-08-08 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | RANKL Employs Distinct Binding Modes to Engage RANK and the Osteoprotegerin Decoy Receptor.
Structure, 20, 2012
|
|
3V0U
 
 | | Crystal Structure of Perakine Reductase, Founder Member of a Novel AKR Subfamily with Unique Conformational Changes during NADPH Binding | | Descriptor: | Perakine reductase | | Authors: | Sun, L, Chen, Y, Rajendran, C, Panjikar, S, Mueller, U, Wang, M, Rosenthal, C, Mindnich, R, Penning, T.M, Stoeckigt, J. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Crystal structure of perakine reductase, founding member of a novel aldo-keto reductase (AKR) subfamily that undergoes unique conformational changes during NADPH binding.
J.Biol.Chem., 287, 2012
|
|
6HY2
 
 | |
3QNN
 
 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dGT Opposite 3tCo | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA Primer, ... | | Authors: | Xia, S, Wang, M, Wang, J, Konigsberg, W.H. | | Deposit date: | 2011-02-08 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Using a Fluorescent Cytosine Analogue tC(o) To Probe the Effect of the Y567 to Ala Substitution on the Preinsertion Steps of dNMP Incorporation by RB69 DNA Polymerase.
Biochemistry, 51, 2012
|
|
