4C5Z
 
 | | Crystal structure of A. niger ochratoxinase | | 分子名称: | OCHRATOXINASE | | 著者 | Dobritzsch, D, Wang, H, Schneider, G, Yu, S. | | 登録日 | 2013-09-17 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural and Functional Characterization of Ochratoxinase, a Novel Mycotoxin Degrading Enzyme.
Biochem.J., 462, 2014
|
|
4KUL
 
 | | Crystal structure of N-terminal acetylated yeast Sir3 BAH domain V83P mutant | | 分子名称: | Regulatory protein SIR3 | | 著者 | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | 登録日 | 2013-05-22 | | 公開日 | 2013-08-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4F7P
 
 | | Crystal Structure of HLA-A*2402 Complexed with a Newly Identified Peptide from 2009H1N1 PB1 (496-505) | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | 著者 | Liu, J, Zhang, S, Tan, S, Yi, Y, Wu, B, Zhu, F, Wang, H, Qi, J, Gao, G.F. | | 登録日 | 2012-05-16 | | 公開日 | 2012-10-10 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Cross-Allele Cytotoxic T Lymphocyte Responses against 2009 Pandemic H1N1 Influenza A Virus among HLA-A24 and HLA-A3 Supertype-Positive Individuals.
J.Virol., 86, 2012
|
|
1VLZ
 
 | |
4FEK
 
 | | Crystal Structure of putative diflavin flavoprotein A 5 (fragment 1-254) from Nostoc sp. PCC 7120, Northeast Structural Genomics Consortium Target NsR435A , Northeast Structural Genomics Consortium (NESG) Target NsR435A | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | 著者 | Kuzin, A, Abashidze, M, Seetharaman, J, Janjua, J, Xiao, R, Foote, E.L, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-05-30 | | 公開日 | 2012-07-11 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Northeast Structural Genomics Consortium Target NsR435A
To be Published
|
|
7DGB
 
 | |
7DGH
 
 | |
7DGF
 
 | |
7DGG
 
 | |
7DGI
 
 | |
7DL4
 
 | |
5YZ0
 
 | | Cryo-EM Structure of human ATR-ATRIP complex | | 分子名称: | ATR-interacting protein, Serine/threonine-protein kinase ATR | | 著者 | Rao, Q, Liu, M, Tian, Y, Wu, Z, Wang, H, Wang, J, Xu, Y. | | 登録日 | 2017-12-11 | | 公開日 | 2018-01-31 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Cryo-EM structure of human ATR-ATRIP complex.
Cell Res., 28, 2018
|
|
4KUI
 
 | | Crystal structure of N-terminal acetylated yeast Sir3 BAH domain | | 分子名称: | ACETIC ACID, ISOPROPYL ALCOHOL, Regulatory protein SIR3 | | 著者 | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | 登録日 | 2013-05-22 | | 公開日 | 2013-08-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JYY
 
 | | Crystal structure of the azide and iron substituted Clostrium difficile SOD2 complex | | 分子名称: | AZIDE ION, FE (III) ION, Superoxide dismutase | | 著者 | Li, W, Ying, T.L, Wang, C.L, Zhao, Y, Wang, H.F, Tan, X.S. | | 登録日 | 2013-04-01 | | 公開日 | 2014-04-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.101 Å) | | 主引用文献 | Crystal structure of the azide and iron substituted Clostrium difficile SOD2 complex
To be Published
|
|
4JZG
 
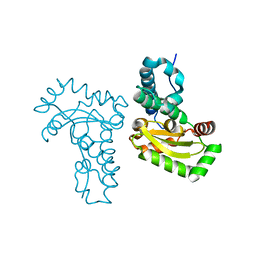 | | Crystal structure of a single cambialistic SOD2 occupied by Manganese ion from Clostridium difficile | | 分子名称: | MANGANESE (II) ION, Superoxide dismutase | | 著者 | Li, W, Wang, C.L, Zhao, Y, Wang, H.F, Tan, S.X. | | 登録日 | 2013-04-02 | | 公開日 | 2014-04-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.321 Å) | | 主引用文献 | Crystal structure of a single cambialistic SOD2 occupied by Manganese ion from Clostridium difficile
To be Published
|
|
4JZ2
 
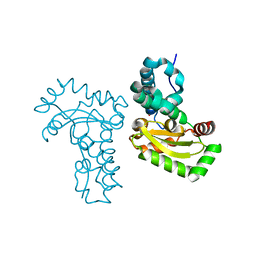 | | Crystal structure of Co ion substituted SOD2 from Clostridium difficile | | 分子名称: | COBALT (II) ION, Superoxide dismutase | | 著者 | Li, W, Ying, T.L, Wang, C.L, Zhao, Y, Wang, H.F, Tan, X.S. | | 登録日 | 2013-04-02 | | 公開日 | 2014-04-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of Co ion substituted SOD2 from Clostridium difficile
To be Published
|
|
3J0A
 
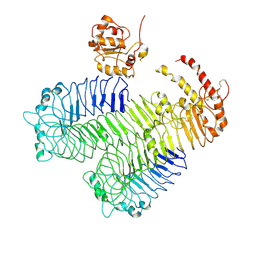 | | Homology model of human Toll-like receptor 5 fitted into an electron microscopy single particle reconstruction | | 分子名称: | Toll-like receptor 5, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Modis, Y, Zhou, K, Kanai, R, Lee, P, Wang, H.W. | | 登録日 | 2011-06-02 | | 公開日 | 2011-12-28 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (26 Å) | | 主引用文献 | Toll-like receptor 5 forms asymmetric dimers in the absence of flagellin.
J.Struct.Biol., 177, 2012
|
|
6JAS
 
 | |
6KH4
 
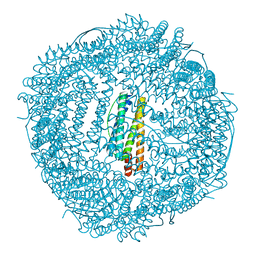 | | Design and crystal structure of protein MOFs with ferritin nanocages as linkers and nickel clusters as nodes | | 分子名称: | FE (III) ION, Ferritin, NICKEL (II) ION | | 著者 | Gu, C, Chen, H, Wang, Y, Zhang, T, Wang, H, Zhao, G. | | 登録日 | 2019-07-12 | | 公開日 | 2020-01-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.302 Å) | | 主引用文献 | Structural Insight into Binary Protein Metal-Organic Frameworks with Ferritin Nanocages as Linkers and Nickel Clusters as Nodes.
Chemistry, 26, 2020
|
|
8K0K
 
 | |
6J15
 
 | | Complex structure of GY-5 Fab and PD-1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GY-5 heavy chain Fab, ... | | 著者 | Chen, D, Tan, S, Zhang, H, Wang, H, Chai, Y, Qi, J, Yan, J, Gao, G.F. | | 登録日 | 2018-12-27 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The FG Loop of PD-1 Serves as a "Hotspot" for Therapeutic Monoclonal Antibodies in Tumor Immune Checkpoint Therapy.
Iscience, 14, 2019
|
|
9BJK
 
 | | Inactive mu opioid receptor bound to Nb6, naloxone and NAM | | 分子名称: | Mu-type opioid receptor, Naloxone, Nalpha-[({(1M)-1-[5-(benzyloxy)pyridin-3-yl]naphthalen-2-yl}sulfanyl)acetyl]-3-methoxy-N,4-dimethyl-L-phenylalaninamide, ... | | 著者 | O'Brien, E.S, Wang, H, Kaavya Krishna, K, Zhang, C, Kobilka, B.K. | | 登録日 | 2024-04-25 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | A mu-opioid receptor modulator that works cooperatively with naloxone.
Nature, 631, 2024
|
|
7EJC
 
 | | human RAD51 presynaptic complex | | 分子名称: | 4-bromanyl-N-(4-bromophenyl)-3-[(phenylmethyl)sulfamoyl]benzamide, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | 著者 | Zhao, L.Y, Xu, J.F, Wang, H.W. | | 登録日 | 2021-04-02 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.97 Å) | | 主引用文献 | Mechanisms of distinctive mismatch tolerance between Rad51 and Dmc1 in homologous recombination.
Nucleic Acids Res., 49, 2021
|
|
7EJ6
 
 | | Yeast Dmc1 presynaptic complex | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), HLJ1_G0016300.mRNA.1.CDS.1, ... | | 著者 | Zhao, L.Y, Xu, J.F, Wang, H.W. | | 登録日 | 2021-04-01 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.21 Å) | | 主引用文献 | Mechanisms of distinctive mismatch tolerance between Rad51 and Dmc1 in homologous recombination.
Nucleic Acids Res., 49, 2021
|
|
6KH5
 
 | | Design and crystal structure of protein MOFs with ferritin nanocages as linkers and nickel clusters as nodes | | 分子名称: | FE (III) ION, Ferritin, NICKEL (II) ION | | 著者 | Gu, C, Chen, H, Wang, Y, Zhang, T, Wang, H, Zhao, G. | | 登録日 | 2019-07-12 | | 公開日 | 2020-01-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.294 Å) | | 主引用文献 | Structural Insight into Binary Protein Metal-Organic Frameworks with Ferritin Nanocages as Linkers and Nickel Clusters as Nodes.
Chemistry, 26, 2020
|
|
