2NTJ
 
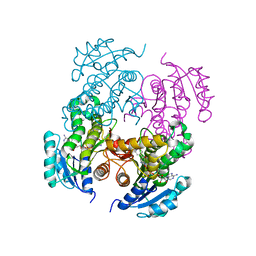 | | Mycobacterium tuberculosis InhA bound with PTH-NAD adduct | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH, {(2R,3S,4R,5R)-5-[(4S)-3-(AMINOCARBONYL)-4-(2-PROPYLISONICOTINOYL)PYRIDIN-1(4H)-YL]-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL}M ETHYL [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | Wang, F, Sacchettini, J.C. | | Deposit date: | 2006-11-07 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of thioamide drug action against tuberculosis and leprosy.
J.Exp.Med., 204, 2007
|
|
2LQI
 
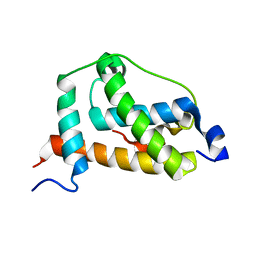 | | NMR structure of FOXO3a transactivation domains (CR2C-CR3) in complex with CBP KIX domain (2l3b conformation) | | Descriptor: | CREB-binding protein, Forkhead box O3 | | Authors: | Wang, F, Marshall, C.B, Yamamoto, K, Li, G.B, Gasmi-Seabrook, G.M.C, Okada, H, Mak, T.W, Ikura, M. | | Deposit date: | 2012-03-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5XSP
 
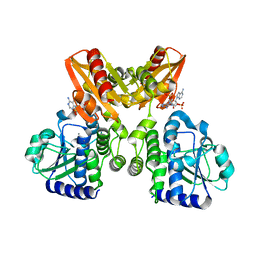 | | The catalytic domain of GdpP with 5'-pApA | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, Phosphodiesterase acting on cyclic dinucleotides | | Authors: | Wang, F, Gu, L. | | Deposit date: | 2017-06-15 | | Release date: | 2018-01-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.146 Å) | | Cite: | Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase
Biochem. J., 475, 2018
|
|
5XT3
 
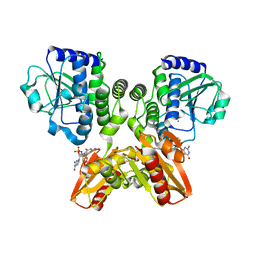 | | The catalytic domain of GdpP with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MANGANESE (II) ION, Phosphodiesterase acting on cyclic dinucleotides | | Authors: | Wang, F, Gu, L. | | Deposit date: | 2017-06-16 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.591 Å) | | Cite: | Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase
Biochem. J., 475, 2018
|
|
7UQK
 
 | |
7UQJ
 
 | | Cryo-EM structure of the S. cerevisiae chromatin remodeler Yta7 hexamer bound to ATPgS and histone H3 tail in state II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase histone chaperone YTA7, Histone H3, ... | | Authors: | Wang, F, Feng, X, Li, H. | | Deposit date: | 2022-04-19 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The Saccharomyces cerevisiae Yta7 ATPase hexamer contains a unique bromodomain tier that functions in nucleosome disassembly.
J.Biol.Chem., 299, 2022
|
|
7UQI
 
 | |
2LQH
 
 | | NMR structure of FOXO3a transactivation domains (CR2C-CR3) in complex with CBP KIX domain (2b3l conformation) | | Descriptor: | CREB-binding protein, Forkhead box O3 | | Authors: | Wang, F, Marshall, C.B, Yamamoto, K, Li, G.B, Gasmi-Seabrook, G.M.C, Okada, H, Mak, T.W, Ikura, M. | | Deposit date: | 2012-03-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5XSI
 
 | | The catalytic domain of GdpP | | Descriptor: | MANGANESE (II) ION, Phosphodiesterase acting on cyclic dinucleotides | | Authors: | Wang, F, Gu, L. | | Deposit date: | 2017-06-14 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase
Biochem. J., 475, 2018
|
|
2NV6
 
 | |
5XSN
 
 | | The catalytic domain of GdpP with c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MANGANESE (II) ION, Phosphodiesterase acting on cyclic dinucleotides | | Authors: | Wang, F, Gu, L. | | Deposit date: | 2017-06-14 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase
Biochem. J., 475, 2018
|
|
5XBW
 
 | | The structure of BrlR | | Descriptor: | Probable transcriptional regulator | | Authors: | Wang, F, Qing, H, Gu, L. | | Deposit date: | 2017-03-21 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.109 Å) | | Cite: | BrlR from Pseudomonas aeruginosa is a receptor for both cyclic di-GMP and pyocyanin.
Nat Commun, 9, 2018
|
|
6ANU
 
 | | Cryo-EM structure of F-actin complexed with the beta-III-spectrin actin-binding domain | | Descriptor: | Actin, cytoplasmic 1, Spectrin beta chain, ... | | Authors: | Wang, F, Orlova, A, Avery, A.W, Hays, T.S, Egelman, E.H. | | Deposit date: | 2017-08-14 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis for high-affinity actin binding revealed by a beta-III-spectrin SCA5 missense mutation.
Nat Commun, 8, 2017
|
|
5XBI
 
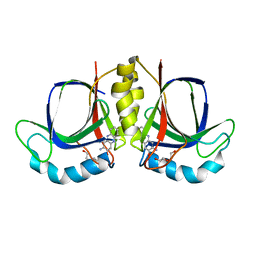 | |
5XBT
 
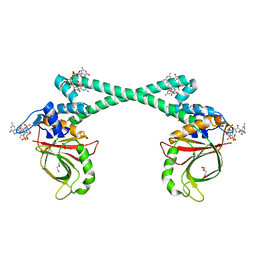 | | The structure of BrlR bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Wang, F, Qing, H, Gu, L. | | Deposit date: | 2017-03-21 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | BrlR from Pseudomonas aeruginosa is a receptor for both cyclic di-GMP and pyocyanin.
Nat Commun, 9, 2018
|
|
7DE0
 
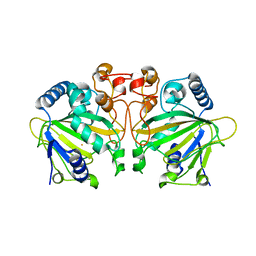 | |
7E0M
 
 | | Crystal structure of phospholipase D | | Descriptor: | Phospholipase, SULFATE ION | | Authors: | Wang, F.H. | | Deposit date: | 2021-01-28 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of a Phospholipase D from the Plant-Associated Bacteria Serratia plymuthica Strain AS9 Reveals a Unique Arrangement of Catalytic Pocket.
Int J Mol Sci, 22, 2021
|
|
7E9V
 
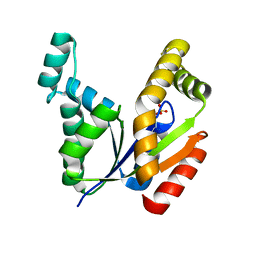 | | The Crystal Structure of human UMP-CMP kinase from Biortus. | | Descriptor: | SULFATE ION, UMP-CMP kinase | | Authors: | Wang, F, Lin, D, Wang, R, Wei, X, Shen, Z, Wang, M. | | Deposit date: | 2021-03-05 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of human UMP-CMP kinase from Biortus.
To Be Published
|
|
7E9W
 
 | | The Crystal Structure of D-psicose-3-epimerase from Biortus. | | Descriptor: | D-psicose 3-epimerase, GLYCEROL, MANGANESE (II) ION | | Authors: | Wang, F, Xu, C, Qi, J, Zhang, M, Tian, F, Wang, M. | | Deposit date: | 2021-03-05 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of D-psicose-3-epimerase from Biortus.
To Be Published
|
|
7ESF
 
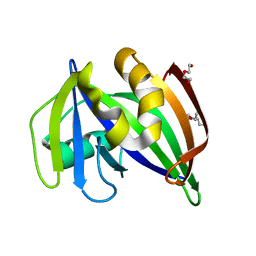 | | The Crystal Structure of human MTH1 from Biortus | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL | | Authors: | Wang, F, Cheng, W, Shang, H, Wang, R, Zhang, B, Tian, F. | | Deposit date: | 2021-05-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Crystal Structure of human MTH1 from Biortus
To Be Published
|
|
7DSF
 
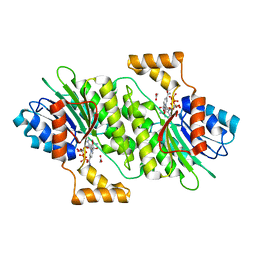 | | The Crystal Structure of human SPR from Biortus. | | Descriptor: | ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Sepiapterin reductase, ... | | Authors: | Wang, F, Lv, Z, Cheng, W, Lin, D, Meng, Q, Zhang, B, Huang, Y. | | Deposit date: | 2020-12-31 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of human SPR from Biortus.
To Be Published
|
|
7DS7
 
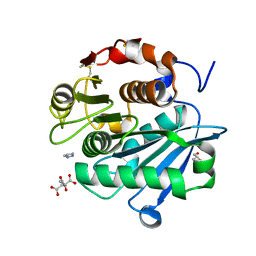 | | The Crystal Structure of Leaf-branch compost cutinase from Biortus. | | Descriptor: | CITRIC ACID, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wang, F, Lv, Z, Cheng, W, Lin, D, Chu, F, Xu, X, Tan, J. | | Deposit date: | 2020-12-30 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal Structure of Leaf-branch compost cutinase from Biortus.
To Be Published
|
|
7D2C
 
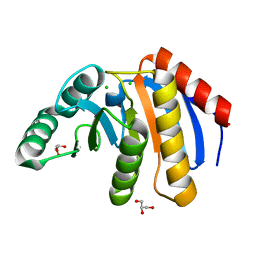 | | The Crystal Structure of human PARP14 from Biortus. | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein mono-ADP-ribosyltransferase PARP14 | | Authors: | Wang, F, Miao, Q, Lv, Z, Cheng, W, Lin, D, Xu, X, Tan, J. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The Crystal Structure of human PARP14 from Biortus.
To Be Published
|
|
7CM2
 
 | | The Crystal Structure of human USP7 USP domain from Biortus | | Descriptor: | GLYCEROL, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Wang, F, Cheng, W, Lv, Z, Lin, D, Zhu, B, Miao, Q, Bao, X, Shang, H. | | Deposit date: | 2020-07-24 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Crystal Structure of human USP7 USP domain from Biortus.
To Be Published
|
|
7CVP
 
 | | The Crystal Structure of human PHGDH from Biortus. | | Descriptor: | D-3-phosphoglycerate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wang, F, Lv, Z, Cheng, W, Lin, D, Miao, Q, Huang, Y. | | Deposit date: | 2020-08-26 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Crystal Structure of human PHGDH from Biortus.
To Be Published
|
|
