6WL8
 
 | | Cryo-EM of Form 2 peptide filament | | Descriptor: | Form 2 peptide | | Authors: | Wang, F, Gnewou, O.M, Xu, C, Su, Z, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-18 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
6WL9
 
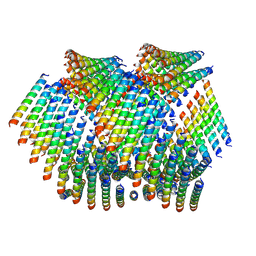 | | Cryo-EM of Form 2 like peptide filament, Form2a | | Descriptor: | peptide Form2a | | Authors: | Wang, F, Beltran, L.C, Gnewou, O.M, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-18 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
7TGG
 
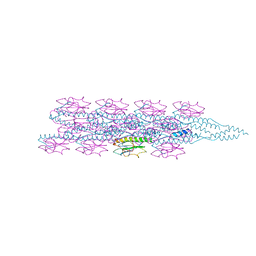 | | Cryo-EM structure of PilA-N and PilA-C from Geobacter sulfurreducens | | Descriptor: | Geopilin domain 1 protein, Geopilin domain 2 protein | | Authors: | Wang, F, Mustafa, K, Chan, C.H, Joshi, K, Bond, D.R, Hochbaum, A.I, Egelman, E.H. | | Deposit date: | 2022-01-07 | | Release date: | 2022-02-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of an extracellular Geobacter OmcE cytochrome filament reveals tetrahaem packing.
Nat Microbiol, 7, 2022
|
|
6WKY
 
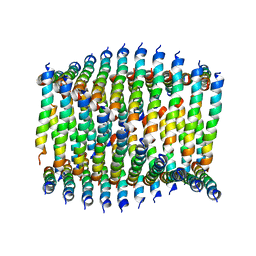 | | Cryo-EM of Form 1 related peptide filament, 29-24-3 | | Descriptor: | peptide 29-24-3 | | Authors: | Wang, F, Gnewou, O.M, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-17 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
7TFS
 
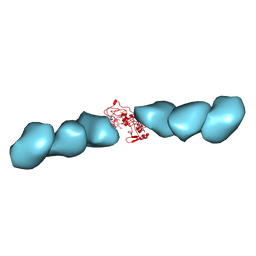 | | Cryo-EM of the OmcE nanowires from Geobacter sulfurreducens | | Descriptor: | Cytochrome c, HEME C | | Authors: | Wang, F, Mustafa, K, Chan, C.H, Joshi, K, Hochbaum, A.I, Bond, D.R, Egelman, E.H. | | Deposit date: | 2022-01-07 | | Release date: | 2022-05-04 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of an extracellular Geobacter OmcE cytochrome filament reveals tetrahaem packing.
Nat Microbiol, 7, 2022
|
|
7TXJ
 
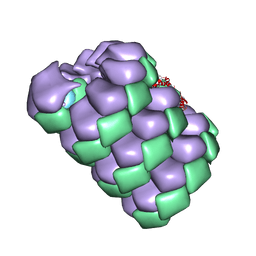 | |
7TXI
 
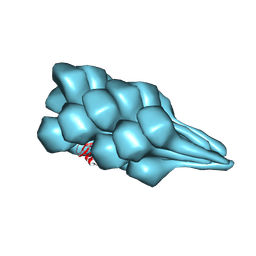 | | Cryo-EM of A. pernix flagellum | | Descriptor: | Probable flagellin 1 | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Baquero, D.P, Krupovic, M, Egelman, E.H. | | Deposit date: | 2022-02-09 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The evolution of archaeal flagellar filaments.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2Z2G
 
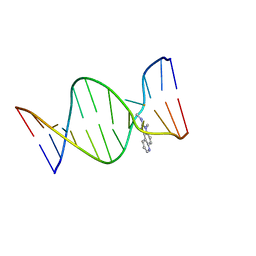 | | NMR Structure of the IQ-modified Dodecamer CTC[IQ]GGCGCCATC | | Descriptor: | 3-METHYL-3H-IMIDAZO[4,5-F]QUINOLIN-2-AMINE, DNA (5'-D(*DCP*DTP*DCP*DGP*DGP*DCP*DGP*DCP*DCP*DAP*DTP*DC)-3'), DNA (5'-D(*DGP*DAP*DTP*DGP*DGP*DCP*DGP*DCP*DCP*DGP*DAP*DG)-3') | | Authors: | Wang, F, Elmquist, C.E, Stover, J.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2007-05-22 | | Release date: | 2007-10-02 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | DNA sequence modulates the conformation of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme
Biochemistry, 46, 2007
|
|
2Z2H
 
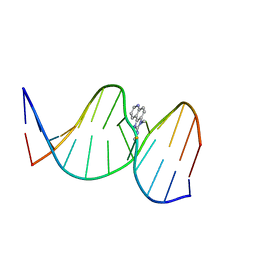 | | NMR Structure of the IQ-modified Dodecamer CTCG[IQ]GCGCCATC | | Descriptor: | 3-METHYL-3H-IMIDAZO[4,5-F]QUINOLIN-2-AMINE, DNA (5'-D(*DCP*DTP*DCP*DGP*DGP*DCP*DGP*DCP*DCP*DAP*DTP*DC)-3'), DNA (5'-D(*DGP*DAP*DTP*DGP*DGP*DCP*DGP*DCP*DCP*DGP*DAP*DG)-3') | | Authors: | Wang, F, Elmquist, C.E, Stover, J.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2007-05-22 | | Release date: | 2007-10-02 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | DNA sequence modulates the conformation of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme
Biochemistry, 46, 2007
|
|
6V7B
 
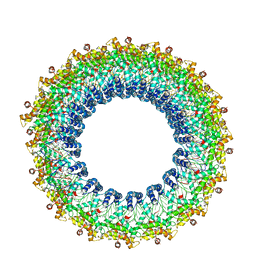 | | Cryo-EM reconstruction of Pyrobaculum filamentous virus 2 (PFV2) | | Descriptor: | A-DNA, Structural protein VP1, Structural protein VP2 | | Authors: | Wang, F, Baquero, D.P, Su, Z, Prangishvili, D, Krupovic, M, Egelman, E.H. | | Deposit date: | 2019-12-08 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of a filamentous virus uncovers familial ties within the archaeal virosphere.
Virus Evol, 6, 2020
|
|
8DST
 
 | |
8FJ5
 
 | | Structure of the Haloferax volcanii archaeal type IV pilus | | Descriptor: | Pilin_N domain-containing protein | | Authors: | Wang, F, Kreutzberger, M.A, Baquero, D.P, Krupovic, M, Egelman, E.H. | | Deposit date: | 2022-12-19 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The evolution of archaeal flagellar filaments.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6NAV
 
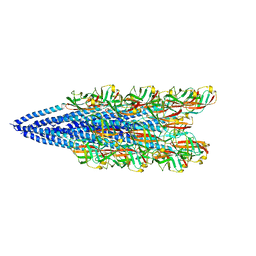 | | Cryo-EM reconstruction of Sulfolobus islandicus LAL14/1 Pilus | | Descriptor: | M9UD72 | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Prangishvili, D, Krupovic, M, Egelman, E.H. | | Deposit date: | 2018-12-06 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | An extensively glycosylated archaeal pilus survives extreme conditions.
Nat Microbiol, 4, 2019
|
|
9B8S
 
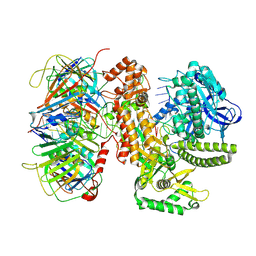 | | Human polymerase epsilon bound to PCNA and DNA in the nucleotide exchange state | | Descriptor: | DNA (5'-D(P*AP*AP*AP*GP*TP*GP*AP*AP*AP*AP*AP*TP*CP*TP*AP*AP*AP*GP*CP*AP*TP*CP*AP*C)-3'), DNA (5'-D(P*GP*TP*GP*AP*TP*GP*CP*TP*TP*TP*AP*GP*AP*TP*TP*TP*TP*TP*C)-3'), DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2024-03-31 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (5.01 Å) | | Cite: | Structures of the human leading strand Pol epsilon-PCNA holoenzyme.
Nat Commun, 15, 2024
|
|
9B8T
 
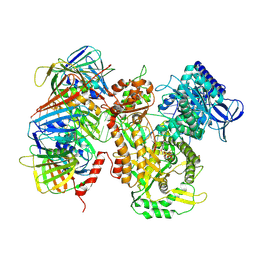 | | Human polymerase epsilon bound to PCNA and DNA in the nucleotide bound state | | Descriptor: | DNA polymerase epsilon catalytic subunit, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2024-03-31 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structures of the human leading strand Pol epsilon-PCNA holoenzyme.
Nat Commun, 15, 2024
|
|
7RX4
 
 | | Cryo-EM reconstruction of AS2 nanotube (Form II like) | | Descriptor: | AS2 peptide | | Authors: | Wang, F, Gnewou, O.M, Solemanifar, A, Xu, C, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2021-08-21 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM of Helical Polymers.
Chem.Rev., 122, 2022
|
|
4ZAS
 
 | | Crystal structure of sugar aminotransferase CalS13 from Micromonospora echinospora | | Descriptor: | CalS13, SULFATE ION, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, F, Singh, S, Miller, M.D, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-04-13 | | Release date: | 2015-04-29 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structure characterization of sugar aminotransferases CalS13 and WecE provides the basis for a unifying structural model for stereochemical outcome.
To Be Published
|
|
2HKC
 
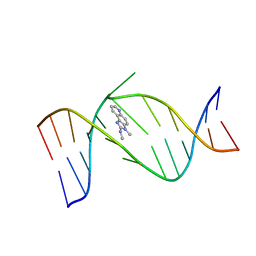 | | NMR Structure of the IQ-modified Dodecamer CTCGGC[IQ]GCCATC | | Descriptor: | 3-METHYL-3H-IMIDAZO[4,5-F]QUINOLIN-2-AMINE, 5'-D(*CP*TP*CP*GP*GP*CP*GP*CP*CP*AP*TP*C)-3', 5'-D(*GP*AP*TP*GP*GP*CP*GP*CP*CP*GP*AP*G)-3' | | Authors: | Wang, F, DeMuro, N.E, Elmquist, C.E, Stover, J.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2006-07-03 | | Release date: | 2006-10-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Base-displaced intercalated structure of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme, a hotspot for -2 bp deletions.
J.Am.Chem.Soc., 128, 2006
|
|
8HC0
 
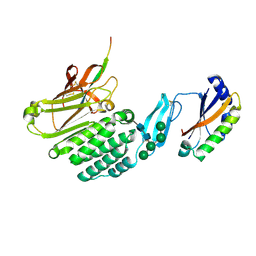 | | Crystal structure of the extracellular domains of GPR110 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G-protein coupled receptor F1, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, F.F, Song, G.J. | | Deposit date: | 2022-11-01 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Extracellular Domains of GPR110.
J.Mol.Biol., 435, 2023
|
|
8WGQ
 
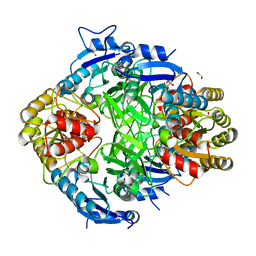 | | The Crystal Structure of L-asparaginase from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, L-asparaginase | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Wang, J. | | Deposit date: | 2023-09-22 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Crystal Structure of L-asparaginase from Biortus.
To Be Published
|
|
8X70
 
 | | The Crystal Structure of IFI16 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Gamma-interferon-inducible protein 16, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Wang, J. | | Deposit date: | 2023-11-22 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of IFI16 from Biortus.
To Be Published
|
|
8XOX
 
 | | The Crystal Structure of FAK2 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, N-methyl-N-{3-[({2-[(2-oxo-2,3-dihydro-1H-indol-5-yl)amino]-5-(trifluoromethyl)pyrimidin-4-yl}amino)methyl]pyridin-2-yl}methanesulfonamide, Protein-tyrosine kinase 2-beta | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Wang, J. | | Deposit date: | 2024-01-02 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of FAK2 from Biortus.
To Be Published
|
|
5D6Y
 
 | | Crystal structure of double tudor domain of human lysine demethylase KDM4A complexed with histone H3K23me3 | | Descriptor: | Lysine-specific demethylase 4A, peptide H3K23me3 (19-28) | | Authors: | Wang, F, Su, Z, Miller, M.D, Denu, J.M, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-08-13 | | Release date: | 2016-02-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Reader domain specificity and lysine demethylase-4 family function.
Nat Commun, 7, 2016
|
|
4ZAH
 
 | | Crystal structure of sugar aminotransferase WecE with External Aldimine VII from Escherichia coli K-12 | | Descriptor: | [[(2R,3S,5R)-5-[5-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3R,4S,5R,6R)-6-methyl-5-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3,4-bis(oxidanyl)oxan-2-yl] hydrogen phosphate, dTDP-4-amino-4,6-dideoxygalactose transaminase | | Authors: | Wang, F, Singh, S, Cao, H, Xu, W, Miller, M.D, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-04-13 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Basis for the Stereochemical Control of Amine Installation in Nucleotide Sugar Aminotransferases.
Acs Chem.Biol., 10, 2015
|
|
8XPN
 
 | | The Crystal Structure of USP8 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ubiquitin carboxyl-terminal hydrolase 8, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Wang, J. | | Deposit date: | 2024-01-04 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of USP8 from Biortus.
To Be Published
|
|
