8K66
 
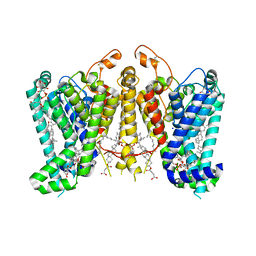 | | Cryo-EM structure of Oryza sativa HKT2;1 at 2.5 angstrom | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Wang, X, Shen, X, Qu, Y, Wang, C, Shen, H. | | Deposit date: | 2023-07-25 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural insights into ion selectivity and transport mechanisms of Oryza sativa HKT2;1 and HKT2;2/1 transporters.
Nat.Plants, 10, 2024
|
|
8JY7
 
 | |
8H0B
 
 | |
8H0A
 
 | |
8H0C
 
 | |
8H09
 
 | |
8H0D
 
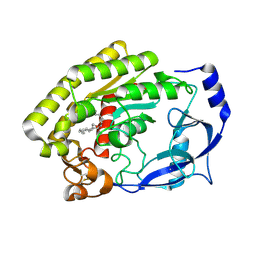 | | Structure of the thermolabile hemolysin from Vibrio alginolyticus (in complex with docosahexaenoic acid) | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, MAGNESIUM ION, ... | | Authors: | Ma, Q, Wang, C. | | Deposit date: | 2022-09-28 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Catalytic site flexibility facilitates the substrate and catalytic promiscuity of Vibrio dual lipase/transferase.
Nat Commun, 14, 2023
|
|
1Z2B
 
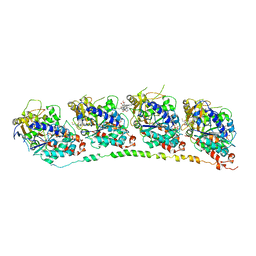 | | Tubulin-colchicine-vinblastine: stathmin-like domain complex | | Descriptor: | (2ALPHA,2'BETA,3BETA,4ALPHA,5BETA)-VINCALEUKOBLASTINE, 2-MERCAPTO-N-[1,2,3,10-TETRAMETHOXY-9-OXO-5,6,7,9-TETRAHYDRO-BENZO[A]HEPTALEN-7-YL]ACETAMIDE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gigant, B, Wang, C, Ravelli, R.B.G, Roussi, F, Steinmetz, M.O, Curmi, P.A, Sobel, A, Knossow, M. | | Deposit date: | 2005-03-08 | | Release date: | 2005-05-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural basis for the regulation of tubulin by vinblastine.
Nature, 435, 2005
|
|
1RJI
 
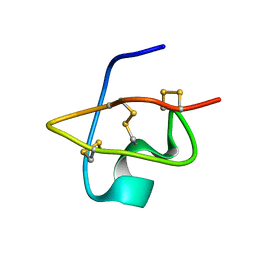 | | Solution Structure of BmKX, a novel potassium channel blocker from the Chinese Scorpion Buthus martensi Karsch | | Descriptor: | potassium channel toxin KX | | Authors: | Cai, Z, Wu, J, Xu, Y, Wang, C.-G, Chi, C.-W, Shi, Y. | | Deposit date: | 2003-11-19 | | Release date: | 2003-12-09 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | A novel short-chain peptide BmKX from the Chinese scorpion Buthus martensi karsch, sequencing, gene cloning and structure determination
Toxicon, 45, 2005
|
|
6KZ6
 
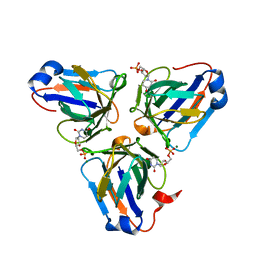 | | Crystal structure of ASFV dUTPase | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, E165R, MAGNESIUM ION | | Authors: | Guo, Y, Chen, C, Li, G.B, Cao, L, Wang, C.W. | | Deposit date: | 2019-09-23 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Structural Insight into African Swine Fever Virus dUTPase Reveals a Novel Folding Pattern in the dUTPase Family.
J.Virol., 94, 2020
|
|
6LCY
 
 | | Crystal structure of Synaptotagmin-7 C2B in complex with IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Synaptotagmin-7 | | Authors: | Zhang, Y, Zhang, X, Rao, F, Wang, C. | | Deposit date: | 2019-11-20 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | 5-IP 7 is a GPCR messenger mediating neural control of synaptotagmin-dependent insulin exocytosis and glucose homeostasis.
Nat Metab, 3, 2021
|
|
5WZH
 
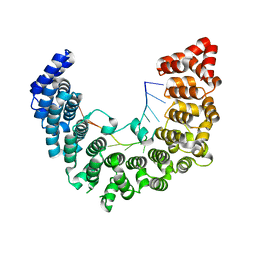 | | Structure of APUM23-GGAAUUGACGG | | Descriptor: | Pumilio homolog 23, RNA (5'-R(*GP*GP*AP*AP*UP*UP*GP*AP*CP*GP*G)-3') | | Authors: | Bao, H, Wang, N, Wang, C, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-01-18 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Structural basis for the specific recognition of 18S rRNA by APUM23.
Nucleic Acids Res., 45, 2017
|
|
5WZI
 
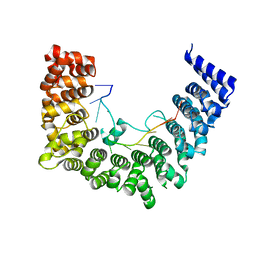 | | Structure of APUM23-GGAGUUGACGG | | Descriptor: | Pumilio homolog 23, RNA (5'-R(*GP*GP*AP*GP*UP*UP*GP*AP*CP*GP*G)-3') | | Authors: | Bao, H, Wang, N, Wang, C, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-01-18 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the specific recognition of 18S rRNA by APUM23.
Nucleic Acids Res., 45, 2017
|
|
8HSY
 
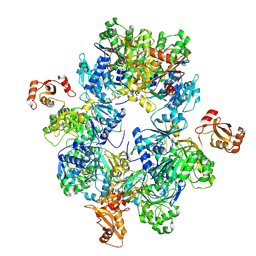 | | Acyl-ACP Synthetase structure | | Descriptor: | Acyl-acyl carrier protein synthetase | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2022-12-20 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Acyl-ACP Synthetase structure
To Be Published
|
|
5XXH
 
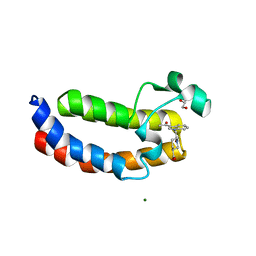 | | Crystal Structure Analysis of the CBP | | Descriptor: | (3S)-1-[2-(3-ethanoylindol-1-yl)ethanoyl]piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, CREB-binding protein, ... | | Authors: | Xiang, Q, Zhang, Y, Wang, C, Song, M. | | Deposit date: | 2017-07-04 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and optimization of 1-(1H-indol-1-yl)ethanone derivatives as CBP/EP300 bromodomain inhibitors for the treatment of castration-resistant prostate cancer.
Eur J Med Chem, 147, 2018
|
|
8JNY
 
 | |
1PYB
 
 | | Crystal Structure of Aquifex aeolicus Trbp111: a Structure-Specific tRNA Binding Protein | | Descriptor: | tRNA-binding protein Trbp111 | | Authors: | Swairjo, M.A, Morales, A.J, Wang, C.C, Ortiz, A.R, Schimmel, P. | | Deposit date: | 2003-07-08 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of trbp111: a structure-specific tRNA-binding protein.
Embo J., 19, 2000
|
|
5Z1T
 
 | | Crystal Structure Analysis of the BRD4(1) | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N-(6-hydroxy-2,2-dimethyl-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-7-yl)-2-methoxybenzene-1-sulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Xiang, Q, Song, M, Wang, C. | | Deposit date: | 2017-12-28 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Y08060: A Selective BET Inhibitor for Treatment of Prostate Cancer.
Acs Med.Chem.Lett., 9, 2018
|
|
4D8O
 
 | |
5WZG
 
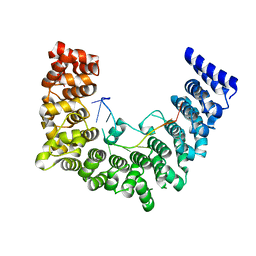 | | Structure of APUM23-GAAUUGACGG | | Descriptor: | Pumilio homolog 23, RNA (5'-R(*GP*AP*AP*UP*UP*GP*AP*CP*GP*G)-3') | | Authors: | Bao, H, Wang, N, Wang, C, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-01-18 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for the specific recognition of 18S rRNA by APUM23.
Nucleic Acids Res., 45, 2017
|
|
2JTC
 
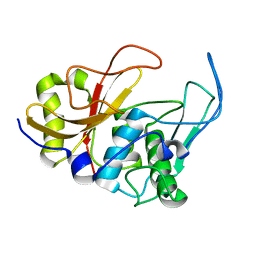 | | 3D structure and backbone dynamics of SPE B | | Descriptor: | Streptopain | | Authors: | Chuang, W, Wang, C, Houng, H, Chen, C, Wang, P. | | Deposit date: | 2007-07-26 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of streptopain: insight into diverse substrate specificity.
J.Biol.Chem., 284, 2009
|
|
5CB5
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | Descriptor: | ACETATE ION, ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase, ... | | Authors: | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
6KKH
 
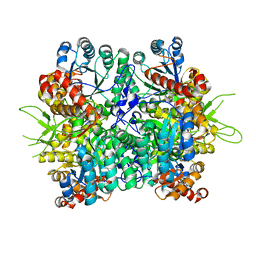 | | Crystal structure of the oxalate bound malyl-CoA lyase from Roseiflexus castenholzii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HpcH/HpaI aldolase, MAGNESIUM ION, ... | | Authors: | Tang, W.R, Wang, Z.G, Zhang, C.Y, Wang, C. | | Deposit date: | 2019-07-25 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The C-terminal domain conformational switch revealed by the crystal structure of malyl-CoA lyase from Roseiflexus castenholzii.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
2ZZQ
 
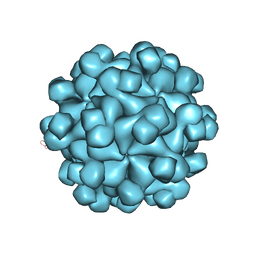 | | Crystal structure analysis of the HEV capsid protein, PORF2 | | Descriptor: | Protein ORF3, Capsid protein | | Authors: | Miyazaki, N, Xing, L, Wang, C.-Y, Li, T.-C, Takeda, N, Higashiura, A, Nakagawa, A, Tsukihara, T, Miyamura, T, Cheng, R.H. | | Deposit date: | 2009-02-23 | | Release date: | 2010-03-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.81 Å) | | Cite: | Role of protein domain-modularity in designating capsid assembly and antigenicity
To be Published
|
|
5CMS
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase, SULFATE ION | | Authors: | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | Deposit date: | 2015-07-17 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
