7XHN
 
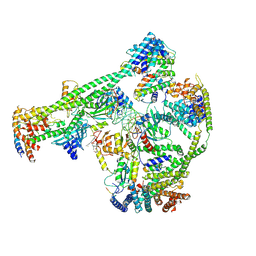 | | Structure of human inner kinetochore CCAN-DNA complex | | Descriptor: | CENP-W, Centromere protein C, Centromere protein H, ... | | Authors: | Sun, L.F, Tian, T, Wang, C.L, Yang, Z.S, Zang, J.Y. | | Deposit date: | 2022-04-09 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural insights into human CCAN complex assembled onto DNA.
Cell Discov, 8, 2022
|
|
8IF5
 
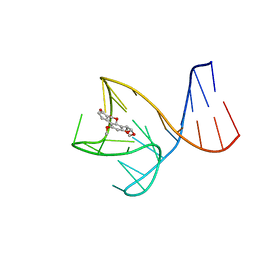 | | AFB1-AF26 APTAMER COMPLEX | | Descriptor: | AFB1 DNA aptamer (26-MER), AFLATOXIN B1 | | Authors: | Xu, G.H, Wang, C, Li, C.G. | | Deposit date: | 2023-02-17 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for high-affinity recognition of aflatoxin B1 by a DNA aptamer.
Nucleic Acids Res., 51, 2023
|
|
4DC2
 
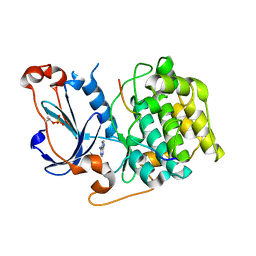 | | Structure of PKC in Complex with a Substrate Peptide from Par-3 | | Descriptor: | ADENINE, Partitioning defective 3 homolog, Protein kinase C iota type | | Authors: | Shang, Y, Wang, C, Yu, J, Zhang, M. | | Deposit date: | 2012-01-17 | | Release date: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Substrate recognition mechanism of atypical protein kinase Cs revealed by the structure of PKC iota in complex with a substrate peptide from Par-3
Structure, 20, 2012
|
|
3S63
 
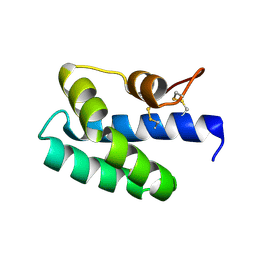 | | Saposin-like protein Na-SLP-1 | | Descriptor: | Saposin-like protein | | Authors: | Willis, C, Wang, C.K, Osman, A, Simon, A, Mulvenna, J, Pickering, D, Riboldi-Tunicliffe, A, Jones, M.K, Loukas, A, Hofmann, A. | | Deposit date: | 2011-05-24 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into the membrane interactions of the saposin-like proteins Na-SLP-1 and Ac-SLP-1 from human and dog hookworm.
Plos One, 6, 2011
|
|
8HSY
 
 | | Acyl-ACP Synthetase structure | | Descriptor: | Acyl-acyl carrier protein synthetase | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2022-12-20 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Acyl-ACP Synthetase structure
To Be Published
|
|
8I22
 
 | | Acyl-ACP synthetase structure bound to pimelic acid monoethyl ester | | Descriptor: | 7-ethoxy-7-oxidanylidene-heptanoic acid, Acyl-acyl carrier protein synthetase | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Acyl-ACP synthetase structure bound to pimelic acid monoethyl ester
To Be Published
|
|
7V1U
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor ZJ12 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(3-ethyl-6-methoxy-1,2-benzoxazol-5-yl)-2-methoxy-benzenesulfonamide | | Authors: | Zhang, M, Wang, C, Zhang, C, Zhang, Y, Xu, Y. | | Deposit date: | 2021-08-06 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Design, synthesis and pharmacological characterization of N-(3-ethylbenzo[d]isoxazol-5-yl) sulfonamide derivatives as BRD4 inhibitors against acute myeloid leukemia.
Acta Pharmacol.Sin., 43, 2022
|
|
8I35
 
 | | Acyl-ACP synthetase structure bound to oleic acid | | Descriptor: | Acyl-acyl carrier protein synthetase, OLEIC ACID | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-16 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Acyl-ACP synthetase structure bound to oleic acid
To Be Published
|
|
8I51
 
 | | Acyl-ACP synthetase structure bound to AMP-MC7 | | Descriptor: | 7-methoxy-7-oxidanylidene-heptanoic acid, ADENOSINE MONOPHOSPHATE, Acyl-acyl carrier protein synthetase, ... | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-21 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Acyl-ACP synthetase structure bound to AMP-MC7
To Be Published
|
|
8I3I
 
 | | Acyl-ACP synthetase structure bound to AMP-PNP in the presence of MgCl2 | | Descriptor: | Acyl-acyl carrier protein synthetase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-17 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Acyl-ACP synthetase structure bound to AMP-PNP in the presence of MgCl2
To Be Published
|
|
8HZX
 
 | | Acyl-ACP synthetase structure-2 | | Descriptor: | Acyl-acyl carrier protein synthetase | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-09 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Acyl-ACP synthetase structure-2
To Be Published
|
|
8I6M
 
 | | Acyl-ACP synthetase structure bound to AMP-C18:1 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acyl-acyl carrier protein synthetase, MAGNESIUM ION, ... | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-28 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Acyl-ACP synthetase structure bound to AMP-C18:1
To Be Published
|
|
8I49
 
 | | Acyl-ACP synthetase structure bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Acyl-acyl carrier protein synthetase | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Acyl-ACP synthetase structure bound to ATP.
To Be Published
|
|
1RZP
 
 | | Crystal Structure of C-Terminal Despentapeptide Nitrite Reductase from Achromobacter Cycloclastes at pH6.2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Li, H.T, Wang, C, Chang, T, Chang, W.C, Liu, M.Y, Le Gall, J, Gui, L.L, Zhang, J.P, An, X.M, Chang, W.R. | | Deposit date: | 2003-12-26 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | pH-profile crystal structure studies of C-terminal despentapeptide nitrite reductase from Achromobacter cycloclastes
Biochem.Biophys.Res.Commun., 316, 2004
|
|
7VHP
 
 | | Structural insights into the membrane microdomain organization by SPFH family proteins | | Descriptor: | ATP-dependent zinc metalloprotease FtsH, Modulator of FtsH protease HflC, Protein HflK | | Authors: | Ma, C.Y, Wang, C.K, Luo, D.Y, Yan, L, Yang, W.X, Li, N.N, Gao, N. | | Deposit date: | 2021-09-22 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural insights into the membrane microdomain organization by SPFH family proteins.
Cell Res., 32, 2022
|
|
7DMG
 
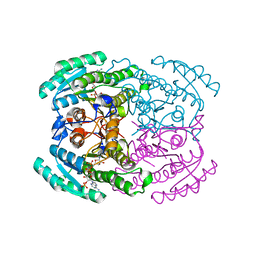 | | Short chain dehydrogenase 2 (SCR2) crystal structure with NADP | | Descriptor: | (S)-specific carbonyl reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2020-12-03 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DLL
 
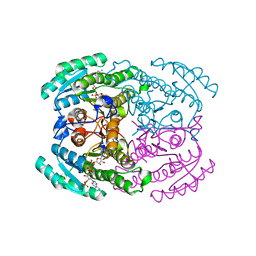 | | Short chain dehydrogenase 2 (SCR2) crystal structure with NADPH | | Descriptor: | (S)-specific carbonyl reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2020-11-28 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
2V7Z
 
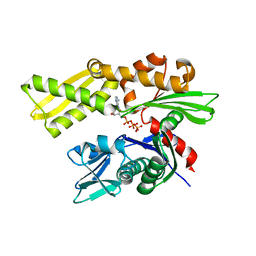 | | Crystal structure of the 70-kDa heat shock cognate protein from Rattus norvegicus in post-ATP hydrolysis state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK COGNATE 71 KDA PROTEIN, PHOSPHATE ION | | Authors: | Chang, Y.-W, Sun, Y.-J, Wang, C, Hsiao, C.-D. | | Deposit date: | 2007-08-02 | | Release date: | 2008-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structures of the 70-kDa Heat Shock Proteins in Domain Disjoining Conformation.
J.Biol.Chem., 283, 2008
|
|
2V7Y
 
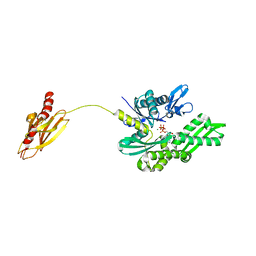 | | Crystal structure of the molecular chaperone DnaK from Geobacillus kaustophilus HTA426 in post-ATP hydrolysis state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHAPERONE PROTEIN DNAK, MAGNESIUM ION, ... | | Authors: | Chang, Y.-W, Sun, Y.-J, Wang, C, Hsiao, C.-D. | | Deposit date: | 2007-08-02 | | Release date: | 2008-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal Structures of the 70-kDa Heat Shock Proteins in Domain Disjoining Conformation.
J.Biol.Chem., 283, 2008
|
|
7EP7
 
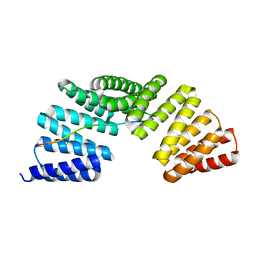 | | The complex structure of Gpsm2 and Whirlin | | Descriptor: | G-protein-signaling modulator 2, Whirlin | | Authors: | Lin, L, Shi, Y, Wang, C, Zhu, J. | | Deposit date: | 2021-04-26 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Promotion of row 1-specific tip complex condensates by Gpsm2-G alpha i provides insights into row identity of the tallest stereocilia.
Sci Adv, 8, 2022
|
|
7ERL
 
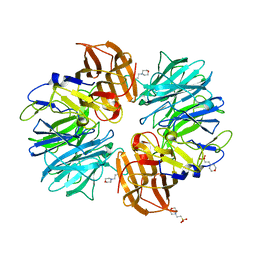 | |
4KEG
 
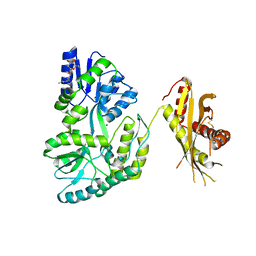 | | Crystal Structure of MBP Fused Human SPLUNC1 | | Descriptor: | MAGNESIUM ION, Maltose-binding periplasmic/Palate lung and nasal epithelium clone fusion protein, octyl beta-D-glucopyranoside | | Authors: | Ning, F, Wang, C, Niu, L, Chu, H.W, Zhang, G. | | Deposit date: | 2013-04-25 | | Release date: | 2014-04-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Lipid Ligands of the SPLUNC1 Protein
To be Published
|
|
1YBJ
 
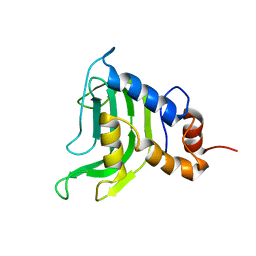 | | Structural and Dynamics studies of both apo and holo forms of the hemophore HasA | | Descriptor: | Hemophore HasA | | Authors: | Wolff, N, Izadi-Pruneyre, N, Couprie, J, Habeck, M, Linge, J, Rieping, W, Wandersman, C, Nilges, M, Delepierre, M, Lecroisey, A. | | Deposit date: | 2004-12-21 | | Release date: | 2005-12-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Comparative analysis of structural and dynamic properties of the loaded and unloaded hemophore HasA: functional implications.
J.Mol.Biol., 376, 2008
|
|
4O8B
 
 | | Crystal structure of transcriptional regulator BswR | | Descriptor: | Uncharacterized protein | | Authors: | Ye, F.Z, Wang, C, Kumar, V, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2013-12-26 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | BswR controls bacterial motility and biofilm formation in Pseudomonas aeruginosa through modulation of the small RNA rsmZ.
Nucleic Acids Res., 42, 2014
|
|
8URY
 
 | | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH bound to ops signal, NusA, mRNA with a 30 nt long spacer, and fMet-tRNA in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S11, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-27 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
