8I03
 
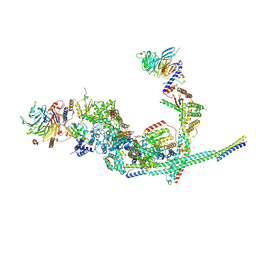 | | Cryo-EM structure of the SIN3L complex from S. pombe | | Descriptor: | Chromatin modification-related protein png2, Histone deacetylase clr6, POTASSIUM ION, ... | | Authors: | Wang, C, Guo, Z, Zhan, X. | | Deposit date: | 2023-01-10 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Two assembly modes for SIN3 histone deacetylase complexes.
Cell Discov, 9, 2023
|
|
5F9H
 
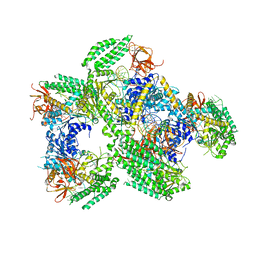 | | Crystal structure of RIG-I helicase-RD in complex with 24-mer 5' triphosphate hairpin RNA | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Probable ATP-dependent RNA helicase DDX58, ... | | Authors: | Wang, C, Marcotrigiano, J, Miller, M, Jiang, F. | | Deposit date: | 2015-12-09 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for m7G recognition and 2'-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4N4W
 
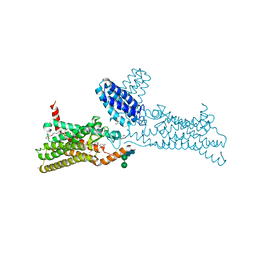 | | Structure of the human smoothened receptor in complex with SANT-1. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (E)-N-(4-benzylpiperazin-1-yl)-1-(3,5-dimethyl-1-phenyl-1H-pyrazol-4-yl)methanimine, Cytochrome b(562),Smoothened homolog, ... | | Authors: | Wang, C, Wu, H, Han, G.W, Cherezov, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs.
Nat Commun, 5, 2014
|
|
5INZ
 
 | | Racemic structure of baboon theta defensin-2 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Theta defensin-2, ... | | Authors: | Wang, C.K, King, G.J, Conibear, A.C, Ramos, M.C, Craik, D.J. | | Deposit date: | 2016-03-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.447 Å) | | Cite: | Mirror Images of Antimicrobial Peptides Provide Reflections on Their Functions and Amyloidogenic Properties.
J.Am.Chem.Soc., 138, 2016
|
|
4IAQ
 
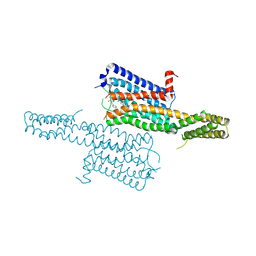 | | Crystal structure of the chimeric protein of 5-HT1B-BRIL in complex with dihydroergotamine (PSI Community Target) | | Descriptor: | Chimera protein of human 5-hydroxytryptamine receptor 1B and E. Coli soluble cytochrome b562, Dihydroergotamine | | Authors: | Wang, C, Jiang, Y, Ma, J, Wu, H, Wacker, D, Katritch, V, Han, G.W, Liu, W, Huang, X, Vardy, E, McCorvy, J.D, Gao, X, Zhou, E.X, Melcher, K, Zhang, C, Bai, F, Yang, H, Yang, L, Jiang, H, Roth, B.L, Cherezov, V, Stevens, R.C, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for molecular recognition at serotonin receptors.
Science, 340, 2013
|
|
4IAR
 
 | | Crystal structure of the chimeric protein of 5-HT1B-BRIL in complex with ergotamine (PSI Community Target) | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera protein of human 5-hydroxytryptamine receptor 1B and E. Coli soluble cytochrome b562, Ergotamine | | Authors: | Wang, C, Jiang, Y, Ma, J, Wu, H, Wacker, D, Katritch, V, Han, G.W, Liu, W, Huang, X, Vardy, E, McCorvy, J.D, Gao, X, Zhou, E.X, Melcher, K, Zhang, C, Bai, F, Yang, H, Yang, L, Jiang, H, Roth, B.L, Cherezov, V, Stevens, R.C, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for molecular recognition at serotonin receptors.
Science, 340, 2013
|
|
7DL9
 
 | | Crystal structure of nucleoside transporter NupG | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Nucleoside permease NupG | | Authors: | Wang, C, Xiao, Q.J, Deng, D. | | Deposit date: | 2020-11-26 | | Release date: | 2021-04-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis for substrate recognition by the bacterial nucleoside transporter NupG.
J.Biol.Chem., 296, 2021
|
|
2KCG
 
 | | Solution structure of cycloviolacin O2 | | Descriptor: | Cycloviolacin-O2 | | Authors: | Wang, C.K. | | Deposit date: | 2008-12-22 | | Release date: | 2009-07-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Despite a conserved cystine knot motif, different cyclotides have different membrane binding modes.
Biophys.J., 97, 2009
|
|
2KCH
 
 | | Solution structure of micelle-bound kalata B2 | | Descriptor: | Kalata-B2 | | Authors: | Wang, C.K. | | Deposit date: | 2008-12-21 | | Release date: | 2009-07-21 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Despite a conserved cystine knot motif, different cyclotides have different membrane binding modes.
Biophys.J., 97, 2009
|
|
7XDG
 
 | | Cryo-EM structures of human mitochondrial NAD(P)+-dependent malic enzyme in a ternary complex with NAD+ and allosteric inhibitor MDSA | | Descriptor: | 5-[(3-carboxy-4-oxidanyl-phenyl)methyl]-2-oxidanyl-benzoic acid, NAD-dependent malic enzyme, mitochondrial, ... | | Authors: | Wang, C.H, Hsieh, J.T, Ho, M.C, Hung, H.C. | | Deposit date: | 2022-03-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Suppression of the human malic enzyme 2 modifies energy metabolism and inhibits cellular respiration
Commun Biol, 6, 2023
|
|
7XDF
 
 | | Cryo-EM structures of human mitochondrial NAD(P)+-dependent malic enzyme in a ternary complex with NAD+ and allosteric inhibitor EA | | Descriptor: | 4-[(3-carboxy-2-oxidanyl-naphthalen-1-yl)methyl]-3-oxidanyl-naphthalene-2-carboxylic acid, NAD-dependent malic enzyme, mitochondrial, ... | | Authors: | Wang, C.H, Hsieh, J.T, Ho, M.C, Hung, H.C. | | Deposit date: | 2022-03-27 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Suppression of the human malic enzyme 2 modifies energy metabolism and inhibits cellular respiration
Commun Biol, 6, 2023
|
|
7XDE
 
 | | Cryo-EM structures of human mitochondrial NAD(P)+-dependent malic enzyme in apo form | | Descriptor: | NAD-dependent malic enzyme, mitochondrial, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wang, C.H, Hsieh, J.T, Ho, M.C, Hung, H.C. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Suppression of the human malic enzyme 2 modifies energy metabolism and inhibits cellular respiration
Commun Biol, 6, 2023
|
|
8JC7
 
 | | Cryo-EM structure of Vibrio campbellii alpha-hemolysin | | Descriptor: | CALCIUM ION, Hemolysin, POTASSIUM ION | | Authors: | Wang, C.H, Yeh, M.K, Ho, M.C, Lin, S.M. | | Deposit date: | 2023-05-10 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.06 Å) | | Cite: | Structural basis for calcium-stimulating pore formation of Vibrio alpha-hemolysin.
Nat Commun, 14, 2023
|
|
8IML
 
 | | Rs2I-Rs2II, Rs1I-Rs1II, RbI-RbII cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster D) | | Descriptor: | CpcA, CpcB, CpcD, ... | | Authors: | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
8IMJ
 
 | | A'1-A'2, A'3-A'4, B1-B2, C1-C2 cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster B) | | Descriptor: | ApcA2, ApcB2, ApcB3, ... | | Authors: | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
8IMN
 
 | | Rt1I-Rt1II, Rt2'I-Rt2'II, Rt3I-Rt3II cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster F) | | Descriptor: | CpcA, CpcB, CpcD, ... | | Authors: | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
8IMM
 
 | | Rs2'I-Rs2'II, Rs1'I-Rs1'II, Rb'I-Rb'II cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster E) | | Descriptor: | CpcA, CpcB, CpcD, ... | | Authors: | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
8IMO
 
 | | Rt1'I-Rt1'II, Rt2I-Rt2II, Rt3'I-Rt3'II cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster G) | | Descriptor: | CpcA, CpcB, CpcD, ... | | Authors: | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
4EVA
 
 | |
2LGG
 
 | | Structure of PHD domain of UHRF1 in complex with H3 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION, histone H3 peptide | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-26 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
8IMI
 
 | | A1-A2, A3-A4, B'1-B'2, C'1-C'2 cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster A) | | Descriptor: | ApcA2, ApcB2, ApcB3, ... | | Authors: | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
8IMK
 
 | | D3-D4, D1-D2, D'3-D'4, D'1-D'2 cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster C) | | Descriptor: | ApcA1, ApcA2, ApcB1, ... | | Authors: | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
2LGK
 
 | | NMR Structure of UHRF1 PHD domains in a complex with histone H3 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION, histone H3 peptide | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-28 | | Release date: | 2011-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
2LGL
 
 | | NMR structure of the UHRF1 PHD domain | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-28 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
3ZH9
 
 | | Bacillus subtilis DNA clamp loader delta protein (YqeN) | | Descriptor: | DELTA, GLYCEROL, SULFATE ION | | Authors: | Suwannachart, C, Sedelnikova, S, Soultanas, P, Oldham, N.J, Rafferty, J.B. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights Into the Structure and Assembly of the Bacillus Subtilis Clamp-Loader Complex and its Interaction with the Replicative Helicase.
Nucleic Acids Res., 41, 2013
|
|
