8FTQ
 
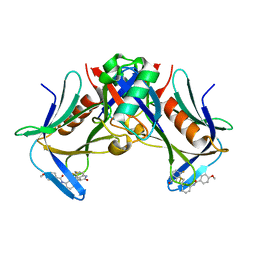 | | Crystal structure of hRpn13 Pru domain in complex with Ubiquitin and XL44 | | Descriptor: | N-(3-{[(3R)-5-fluoro-2-oxo-2,3-dihydro-1H-indol-3-yl]methyl}phenyl)-4-methoxybenzamide, Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Walters, K.J, Lu, X, Chandravanshi, M. | | Deposit date: | 2023-01-13 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structure-based designed small molecule depletes hRpn13 Pru and a select group of KEN box proteins.
Nat Commun, 15, 2024
|
|
1J8C
 
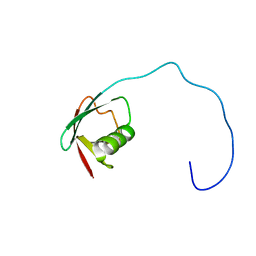 | | Solution Structure of the Ubiquitin-like Domain of hPLIC-2 | | Descriptor: | ubiquitin-like protein hPLIC-2 | | Authors: | Walters, K.J, Kleijnen, M.F, Goh, A.M, Wagner, G, Howley, P.M. | | Deposit date: | 2001-05-21 | | Release date: | 2002-03-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a.
Biochemistry, 41, 2002
|
|
1AJY
 
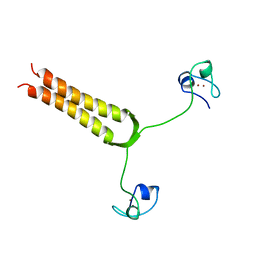 | | STRUCTURE AND MOBILITY OF THE PUT3 DIMER: A DNA PINCER, NMR, 13 STRUCTURES | | Descriptor: | PUT3, ZINC ION | | Authors: | Walters, K.J, Dayie, K.T, Reece, R.J, Ptashne, M, Wagner, G. | | Deposit date: | 1997-05-12 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and mobility of the PUT3 dimer.
Nat.Struct.Biol., 4, 1997
|
|
1CKV
 
 | |
1OQY
 
 | | Structure of the DNA repair protein hHR23a | | Descriptor: | UV excision repair protein RAD23 homolog A | | Authors: | Walters, K.J, Lech, P.J, Goh, A.M, Wang, Q, Howley, P.M. | | Deposit date: | 2003-03-11 | | Release date: | 2003-10-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | DNA-repair protein hHR23a alters its protein structure upon binding proteasomal subunit S5a
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1QZE
 
 | | HHR23a protein structure based on residual dipolar coupling data | | Descriptor: | UV excision repair protein RAD23 homolog A | | Authors: | Walters, K.J, Lech, P.J, Goh, A.M, Wang, Q, Howley, P.M. | | Deposit date: | 2003-09-16 | | Release date: | 2003-10-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | DNA-repair protein hHR23a alters its protein structure upon binding proteasomal subunit S5a
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
8GI6
 
 | | Crystal structure of RhoA mutant L69R complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | Authors: | Chen, X, Qian, X, Chandravanshi, M, Lowy, D.R, Walters, K.J. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Ras-like GTPases mutant structures
To be published
|
|
8GI3
 
 | | Crystal structure of RhoA mutant L69P complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | Authors: | Chen, X, Qian, X, Chandravanshi, M, Lowy, D.R, Walters, K.J. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Ras-like GTPases mutants structure
To be published
|
|
6CO4
 
 | |
2Z59
 
 | | Complex Structures of Mouse Rpn13 (22-130aa) and ubiquitin | | Descriptor: | Protein ADRM1, Ubiquitin | | Authors: | Chen, X, Schreiner, P, Groll, M, Walters, K.J. | | Deposit date: | 2007-07-01 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction.
Nature, 453, 2008
|
|
2Z4D
 
 | |
5W77
 
 | | Solution structure of the MYC G-quadruplex bound to small molecule DC-34 | | Descriptor: | 4-[(azepan-1-yl)methyl]-5-hydroxy-2-methyl-N-[4-(trifluoromethyl)phenyl]-1-benzofuran-3-carboxamide, DNA (5'-D(*TP*GP*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*A)-3'), POTASSIUM ION | | Authors: | Chen, X, Walters, K.J. | | Deposit date: | 2017-06-19 | | Release date: | 2018-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Chemical and structural studies provide a mechanistic basis for recognition of the MYC G-quadruplex.
Nat Commun, 9, 2018
|
|
6E5N
 
 | |
6MUN
 
 | | Structure of hRpn10 bound to UBQLN2 UBL | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4, Ubiquilin-2 | | Authors: | Chen, X, Walters, K.J. | | Deposit date: | 2018-10-23 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of hRpn10 Bound to UBQLN2 UBL Illustrates Basis for Complementarity between Shuttle Factors and Substrates at the Proteasome.
J.Mol.Biol., 431, 2019
|
|
2N3T
 
 | |
2N3W
 
 | |
2N3U
 
 | |
1YX4
 
 | |
2NBW
 
 | | Solution structure of the Rpn1 T1 site with the Rad23 UBL domain | | Descriptor: | 26S proteasome regulatory subunit RPN1, UV excision repair protein RAD23 | | Authors: | Chen, X, Walters, K.J. | | Deposit date: | 2016-03-14 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome.
Structure, 24, 2016
|
|
1YX6
 
 | |
2NBV
 
 | |
1YX5
 
 | |
2NBU
 
 | |
2N3V
 
 | |
6UYJ
 
 | | hRpn13:hRpn2:K48-diubiquitin | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Lu, X, Walters, K.J. | | Deposit date: | 2019-11-13 | | Release date: | 2020-03-11 | | Last modified: | 2020-05-20 | | Method: | SOLUTION NMR | | Cite: | An Extended Conformation for K48 Ubiquitin Chains Revealed by the hRpn2:Rpn13:K48-Diubiquitin Structure.
Structure, 28, 2020
|
|
