1JMK
 
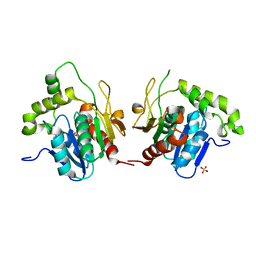 | | Structural Basis for the Cyclization of the Lipopeptide Antibiotic Surfactin by the Thioesterase Domain SrfTE | | Descriptor: | SULFATE ION, Surfactin Synthetase | | Authors: | Bruner, S.D, Weber, T, Kohli, R.M, Schwarzer, D, Marahiel, M.A, Walsh, C.T, Stubbs, M.T. | | Deposit date: | 2001-07-18 | | Release date: | 2002-03-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis for the cyclization of the lipopeptide antibiotic surfactin by the thioesterase domain SrfTE.
Structure, 10, 2002
|
|
1RRV
 
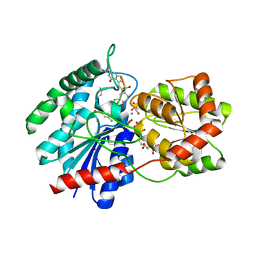 | | X-ray crystal structure of TDP-vancosaminyltransferase GtfD as a complex with TDP and the natural substrate, desvancosaminyl vancomycin. | | Descriptor: | DESVANCOSAMINYL VANCOMYCIN, GLYCEROL, GLYCOSYLTRANSFERASE GTFD, ... | | Authors: | Mulichak, A.M, Lu, W, Losey, H.C, Walsh, C.T, Garavito, R.M. | | Deposit date: | 2003-12-09 | | Release date: | 2004-05-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Vancosaminyltransferase Gtfd from the Vancomycin Biosynthetic Pathway: Interactions with Acceptor and Nucleotide Ligands
Biochemistry, 43, 2004
|
|
2GRT
 
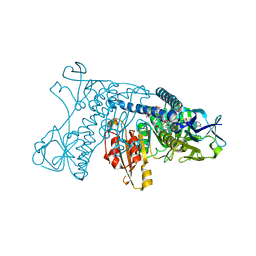 | | HUMAN GLUTATHIONE REDUCTASE A34E, R37W MUTANT, OXIDIZED GLUTATHIONE COMPLEX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE, OXIDIZED GLUTATHIONE DISULFIDE | | Authors: | Stoll, V.S, Simpson, S.J, Krauth-Siegel, R.L, Walsh, C.T, Pai, E.F. | | Deposit date: | 1997-02-12 | | Release date: | 1997-08-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Glutathione reductase turned into trypanothione reductase: structural analysis of an engineered change in substrate specificity.
Biochemistry, 36, 1997
|
|
1CSA
 
 | |
4EIP
 
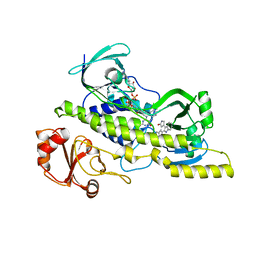 | | Native and K252c bound RebC-10x | | Descriptor: | 6,7,12,13-tetrahydro-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazol-5-one, FLAVIN-ADENINE DINUCLEOTIDE, Putative FAD-monooxygenase | | Authors: | Goldman, P.J, Ryan, K.S, Howard-Jones, A.R, Hamill, M.J, Elliott, S.J, Walsh, C.T, Drennan, C.L. | | Deposit date: | 2012-04-05 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.332 Å) | | Cite: | An Unusual Role for a Mobile Flavin in StaC-like Indolocarbazole Biosynthetic Enzymes.
Chem.Biol., 19, 2012
|
|
2LIU
 
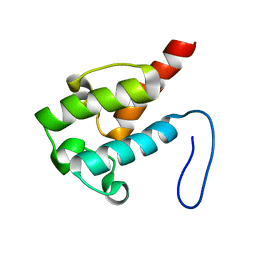 | | NMR structure of holo-ACPI domain from CurA module from Lyngbya majuscula | | Descriptor: | CurA | | Authors: | Busche, A.E, Gottstein, D, Hein, C, Ripin, N, Pader, I, Tufar, P, Eisman, E.B, Gu, L, Walsh, C.T, Loehr, F, Sherman, D.H, Guntert, P, Dotsch, V. | | Deposit date: | 2011-09-01 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of Molecular Interactions between ACP and Halogenase Domains in the Curacin A Polyketide Synthase.
Acs Chem.Biol., 7, 2012
|
|
2MLP
 
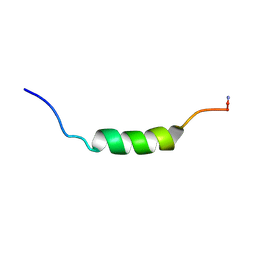 | | MICROCIN LEADER PEPTIDE FROM E. COLI, NMR, 25 STRUCTURES | | Descriptor: | MCBA PROPEPTIDE | | Authors: | Kim, S, Sinha Roy, R, Walsh, C.T, Baleja, J.D. | | Deposit date: | 1998-01-21 | | Release date: | 1998-07-22 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Role of the microcin B17 propeptide in substrate recognition: solution structure and mutational analysis of McbA1-26.
Chem.Biol., 5, 1998
|
|
2ISJ
 
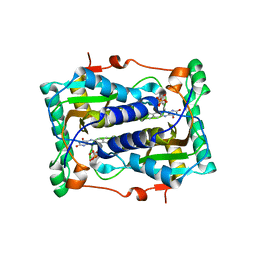 | | BluB bound to oxidized FMN | | Descriptor: | BluB, FLAVIN MONONUCLEOTIDE | | Authors: | Larsen, N.A, Taga, M.E, Howard-Jones, A.R, Walsh, C.T, Walker, G.C. | | Deposit date: | 2006-10-17 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | BluB cannibalizes flavin to form the lower ligand of vitamin B12.
Nature, 446, 2007
|
|
2LIW
 
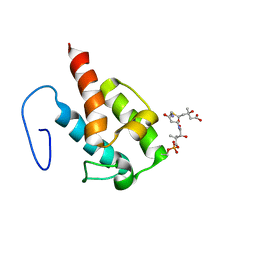 | | NMR structure of HMG-ACPI domain from CurA module from Lyngbya majuscula | | Descriptor: | 3-HYDROXY-3-METHYL-GLUTARIC ACID, 4'-PHOSPHOPANTETHEINE, CurA | | Authors: | Busche, A.E, Gottstein, D, Hein, C, Ripin, N, Pader, I, Tufar, P, Eisman, E.B, Gu, L, Walsh, C.T, Loehr, F, Sherman, D.H, Guntert, P, Dotsch, V. | | Deposit date: | 2011-09-01 | | Release date: | 2011-12-21 | | Last modified: | 2012-03-14 | | Method: | SOLUTION NMR | | Cite: | Characterization of Molecular Interactions between ACP and Halogenase Domains in the Curacin A Polyketide Synthase.
Acs Chem.Biol., 7, 2012
|
|
2ISL
 
 | | BluB bound to reduced flavin (FMNH2) and molecular oxygen. (clear crystal form) | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, BluB, OXYGEN MOLECULE | | Authors: | Larsen, N.A, Taga, M.E, Howard-Jones, A.R, Walsh, C.T, Walker, G.C. | | Deposit date: | 2006-10-17 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | BluB cannibalizes flavin to form the lower ligand of vitamin B12.
Nature, 446, 2007
|
|
2ISK
 
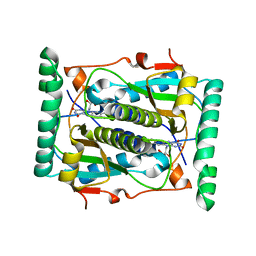 | | BluB bound to flavin anion (charge transfer complex) | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, BluB | | Authors: | Larsen, N.A, Taga, M.E, Howard-Jones, A.R, Walsh, C.T, Walker, G.C. | | Deposit date: | 2006-10-17 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | BluB cannibalizes flavin to form the lower ligand of vitamin B12.
Nature, 446, 2007
|
|
2GZR
 
 | |
2GZS
 
 | |
2K2Q
 
 | | complex structure of the external thioesterase of the Surfactin-synthetase with a carrier domain | | Descriptor: | Surfactin synthetase thioesterase subunit, Tyrocidine synthetase 3 (Tyrocidine synthetase III) | | Authors: | Koglin, A, Lohr, F, Bernhard, F, Rogov, V.V, Frueh, D.P, Strieter, E.R, Mofid, M.R, Guntert, P, Wagner, G, Walsh, C.T, Marahiel, M.A, Dotsch, V. | | Deposit date: | 2008-04-10 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the selectivity of the external thioesterase of the surfactin synthetase.
Nature, 454, 2008
|
|
1L5A
 
 | | Crystal Structure of VibH, an NRPS Condensation Enzyme | | Descriptor: | amide synthase | | Authors: | Keating, T.A, Marshall, C.G, Walsh, C.T, Keating, A.E. | | Deposit date: | 2002-03-06 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The structure of VibH represents nonribosomal peptide synthetase condensation, cyclization and epimerization domains.
Nat.Struct.Biol., 9, 2002
|
|
