4W6K
 
 | |
8DJ8
 
 | | Crystal Structure of Calgreen 1 protein | | Descriptor: | Calgreen One | | Authors: | Hung, L.-W, Nguyen, H.B, Waldo, G. | | Deposit date: | 2022-06-30 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of Engineered Variants of GFP-type Green Fluorescent protein from Corynactis californica (tentative)
To Be Published
|
|
3MML
 
 | | Allophanate Hydrolase Complex from Mycobacterium smegmatis, Msmeg0435-Msmeg0436 | | Descriptor: | Allophanate hydrolase subunit 1, Allophanate hydrolase subunit 2, CHLORIDE ION | | Authors: | Kaufmann, M, Chernishof, I, Shin, A, Germano, D, Sawaya, M.R, Waldo, G.S, Arbing, M.A, Perry, J, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-04-20 | | Release date: | 2010-04-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Allphanate Hydrolase Complex from M. smegmatis, Msmeg0435-Msmeg0436
To be Published
|
|
1BKB
 
 | | INITIATION FACTOR 5A FROM ARCHEBACTERIUM PYROBACULUM AEROPHILUM | | Descriptor: | TRANSLATION INITIATION FACTOR 5A | | Authors: | Peat, T.S, Newman, J, Waldo, G.S, Berendzen, J, Terwilliger, T.C. | | Deposit date: | 1998-07-05 | | Release date: | 1998-11-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of translation initiation factor 5A from Pyrobaculum aerophilum at 1.75 A resolution.
Structure, 6, 1998
|
|
8U23
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP 8 | | Descriptor: | Green Fluorescent Protein Variant #8, ccGFP 8 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
6WZN
 
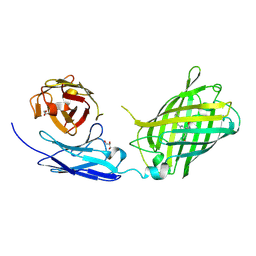 | | Crystal Structure of a Fluorescent Single Chain Fv Chimera | | Descriptor: | Fluorescent Single Chain Fv Chimera, GLYCEROL | | Authors: | Close, D, Velappan, N, Hung, L.W, Naranjo, L, Hemez, C, DeVore, N, McCullough, D, Lillo, A.M, Waldo, G, Bradbury, A.R.M. | | Deposit date: | 2020-05-14 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Construction, characterization and crystal structure of a fluorescent single-chain Fv chimera.
Protein Eng.Des.Sel., 34, 2021
|
|
4W6S
 
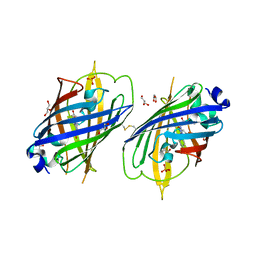 | | Crystal Structure of Full-Length Split GFP Mutant K126C Disulfide Dimer, P 43 21 2 Space Group | | Descriptor: | GLYCEROL, PHOSPHATE ION, fluorescent protein E124H/K126C | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-20 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W7A
 
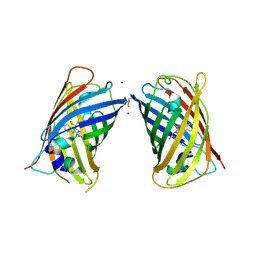 | | Crystal Structure of Full-Length Split GFP Mutant D21H/K26C Disulfide and Metal-Mediated Dimer, P 21 21 21 Space Group, Form 4 | | Descriptor: | COPPER (II) ION, fluorescent protein D21H/K26C | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-21 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.603 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W6M
 
 | |
4W6U
 
 | | Crystal Structure of Full-Length Split GFP Mutant E115H/T118H With Nickel Mediated Crystal Contacts, P 21 21 21 Space Group | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, NICKEL (II) ION, ... | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-20 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W7D
 
 | | Crystal Structure of Full-Length Split GFP Mutant D21H/K26H With Copper Mediated Crystal Contacts, P 21 21 21 Space Group | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, COPPER (II) ION, GLYCEROL, ... | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-21 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
1JAD
 
 | | C-terminal Domain of Turkey PLC-beta | | Descriptor: | SULFATE ION, phospholipase C beta | | Authors: | Singer, A.U, Waldo, G.L, Harden, T.K, Sondek, J. | | Deposit date: | 2001-05-30 | | Release date: | 2001-12-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A unique fold of phospholipase C-beta mediates dimerization and interaction with G alpha q.
Nat.Struct.Biol., 9, 2002
|
|
4W75
 
 | | Crystal Structure of Full-Length Split GFP Mutant D21H/K26C Disulfide and Metal-Mediated Dimer, P 21 21 21 Space Group, Form 1 | | Descriptor: | COPPER (II) ION, fluorescent protein D21H/K26C | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-21 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.473 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W69
 
 | |
4W6D
 
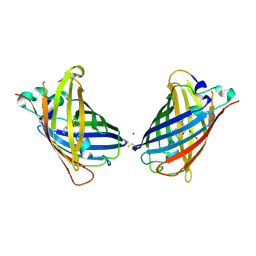 | | Crystal Structure of Full-Length Split GFP Mutant K26C Disulfide Dimer, P 32 2 1 Space Group, Form 1 | | Descriptor: | MAGNESIUM ION, fluorescent protein K26C | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-20 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W6I
 
 | |
4W77
 
 | | Crystal Structure of Full-Length Split GFP Mutant D21H/K26C Disulfide and Metal-Mediated Dimer, P 21 21 21 Space Group, Form 3 | | Descriptor: | COPPER (II) ION, fluorescent protein D21H/K26C | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-21 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
8U20
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP 5 | | Descriptor: | Green Fluorescent Protein Variant #5, ccGFP 5 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
8U22
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP 7 | | Descriptor: | Green Fluorescent Protein Variant #7, ccGFP 7 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
8U21
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP E6 | | Descriptor: | Green Fluorescent Protein Variant E6, ccGFP E6 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
8U24
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP 9 | | Descriptor: | Green Fluorescent Protein Variant #9, ccGFP 9 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
1XQI
 
 | | Crystal Structure Analysis of an NDP kinase from Pyrobaculum aerophilum | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Nucleoside diphosphate kinase, TRIETHYLENE GLYCOL | | Authors: | Pedelacq, J.D, Waldo, G.S, Cabantous, S, Liong, E.C, Berendzen, J, Terwilliger, T.C. | | Deposit date: | 2004-10-12 | | Release date: | 2005-09-20 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional features of an NDP kinase from the hyperthermophile crenarchaeon Pyrobaculum aerophilum
Protein Sci., 14, 2005
|
|
2B3P
 
 | | Crystal structure of a superfolder green fluorescent protein | | Descriptor: | ACETIC ACID, CADMIUM ION, green fluorescent protein | | Authors: | Pedelacq, J.D, Cabantous, S, Tran, T.H, Terwilliger, T.C, Waldo, G.S. | | Deposit date: | 2005-09-20 | | Release date: | 2005-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineering and characterization of a superfolder green fluorescent protein.
Nat.Biotechnol., 24, 2006
|
|
2B3Q
 
 | | Crystal structure of a well-folded variant of green fluorescent protein | | Descriptor: | MAGNESIUM ION, green fluorescent protein | | Authors: | Pedelacq, J.D, Cabantous, S, Tran, T.H, Terwilliger, T.C, Waldo, G.S. | | Deposit date: | 2005-09-20 | | Release date: | 2005-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Engineering and characterization of a superfolder green fluorescent protein.
Nat.Biotechnol., 24, 2006
|
|
4KF4
 
 | | Crystal Structure of sfCherry | | Descriptor: | fluorescent protein sfCherry | | Authors: | Nguyen, H.B, Hung, L.-W, Yeates, T.O, Waldo, G.S, Terwilliger, T.C. | | Deposit date: | 2013-04-26 | | Release date: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Split green fluorescent protein as a modular binding partner for protein crystallization.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
