3WLU
 
 | | Crystal Structure of human galectin-9 NCRD with Selenolactose | | Descriptor: | 2-(trimethylsilyl)ethyl 4-O-beta-D-galactopyranosyl-6-Se-methyl-6-seleno-beta-D-glucopyranoside, Galectin-9 | | Authors: | Makyio, H, Suzuki, T, Ando, H, Yamada, Y, Ishida, H, Kiso, M, Wakatsuki, S, Kato, R. | | Deposit date: | 2013-11-14 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Expanded potential of seleno-carbohydrates as a molecular tool for X-ray structural determination of a carbohydrate-protein complex with single/multi-wavelength anomalous dispersion phasing
Bioorg.Med.Chem., 22, 2014
|
|
3VHT
 
 | | Crystal structure of GFP-Wrnip1 UBZ domain fusion protein in complex with ubiquitin | | Descriptor: | Green fluorescent protein, Green fluorescent protein,ATPase WRNIP1, Ubiquitin, ... | | Authors: | Suzuki, N, Wakatsuki, S, Kawasaki, M. | | Deposit date: | 2011-09-06 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel mode of ubiquitin recognition by the ubiquitin-binding zinc finger domain of WRNIP1.
Febs J., 2016
|
|
3ALB
 
 | | Cyclic Lys48-linked tetraubiquitin | | Descriptor: | SULFATE ION, ubiquitin | | Authors: | Satoh, T, Sakata, E, Yamamoto, S, Yamaguchi, Y, Sumiyoshi, A, Wakatsuki, S, Kato, K. | | Deposit date: | 2010-07-29 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of cyclic Lys48-linked tetraubiquitin
Biochem.Biophys.Res.Commun., 2010
|
|
3AI4
 
 | | Crystal structure of yeast enhanced green fluorescent protein - mouse polymerase iota ubiquitin binding motif fusion protein | | Descriptor: | SULFATE ION, yeast enhanced green fluorescent protein,DNA polymerase iota | | Authors: | Suzuki, N, Wakatsuki, S, Kawasaki, M. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallization of small proteins assisted by green fluorescent protein
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3AI5
 
 | | Crystal structure of yeast enhanced green fluorescent protein-ubiquitin fusion protein | | Descriptor: | 1,2-ETHANEDIOL, yeast enhanced green fluorescent protein,Ubiquitin | | Authors: | Suzuki, N, Wakatsuki, S, Kawasaki, M. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallization of small proteins assisted by green fluorescent protein
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3BC1
 
 | | Crystal Structure of the complex Rab27a-Slp2a | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-27A, ... | | Authors: | Chavas, L.M.G, Ihara, K, Kawasaki, M, Wakatsuki, S. | | Deposit date: | 2007-11-12 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Elucidation of Rab27 recruitment by its effectors: structure of Rab27a bound to Exophilin4/Slp2-a
Structure, 16, 2008
|
|
2D7I
 
 | | Crystal structure of pp-GalNAc-T10 with UDP, GalNAc and Mn2+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kubota, T, Shiba, T, Sugioka, S, Kato, R, Wakatsuki, S, Narimatsu, H. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of carbohydrate transfer activity by human UDP-GalNAc: polypeptide alpha-N-acetylgalactosaminyltransferase (pp-GalNAc-T10)
J.Mol.Biol., 359, 2006
|
|
2D7R
 
 | | Crystal structure of pp-GalNAc-T10 complexed with GalNAc-Ser on lectin domain | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kubota, T, Shiba, T, Sugioka, S, Kato, R, Wakatsuki, S, Narimatsu, H. | | Deposit date: | 2005-11-25 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of carbohydrate transfer activity by human UDP-GalNAc: polypeptide alpha-N-acetylgalactosaminyltransferase (pp-GalNAc-T10)
J.Mol.Biol., 359, 2006
|
|
2D0J
 
 | | Crystal Structure of Human GlcAT-S Apo Form | | Descriptor: | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 2 | | Authors: | Shiba, T, Kakuda, S, Ishiguro, M, Oka, S, Kawasaki, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2005-08-03 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of GlcAT-S, a human glucuronyltransferase, involved in the biosynthesis of the HNK-1 carbohydrate epitope
To be published
|
|
2ZAO
 
 | | Crystal structure of mouse SKD1/VPS4B ADP-form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Vacuolar protein sorting-associating protein 4B | | Authors: | Inoue, M, Kawasaki, M, Kamikubo, H, Kataoka, M, Kato, R, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2007-10-08 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Nucleotide-dependent conformational changes and assembly of the AAA ATPase SKD1/VPS4B
Traffic, 9, 2008
|
|
2ZN9
 
 | | Crystal structure of Ca2+-bound form of des3-20ALG-2 | | Descriptor: | CALCIUM ION, DODECAETHYLENE GLYCOL, NONAETHYLENE GLYCOL, ... | | Authors: | Suzuki, H, Kawasaki, M, Inuzuka, T, Kakiuchi, T, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Ca(2+)-Dependent Formation of ALG-2/Alix Peptide Complex: Ca(2+)/EF3-Driven Arginine Switch Mechanism
Structure, 16, 2008
|
|
2ZHK
 
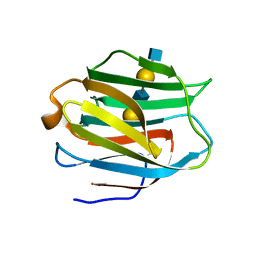 | | Crystal structure of human galectin-9 N-terminal CRD in complex with N-acetyllactosamine dimer (crystal 1) | | Descriptor: | Galectin-9, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Nagae, M, Nishi, N, Murata, T, Usui, T, Nakamura, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2008-02-06 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the recognition mechanism of poly-N-acetyllactosamine by the human galectin-9 N-terminal carbohydrate recognition domain.
Glycobiology, 19, 2009
|
|
2ZHL
 
 | | Crystal structure of human galectin-9 N-terminal CRD in complex with N-acetyllactosamine dimer (crystal 2) | | Descriptor: | Galectin-9, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Nagae, M, Nishi, N, Murata, T, Usui, T, Nakamura, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2008-02-06 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of the recognition mechanism of poly-N-acetyllactosamine by the human galectin-9 N-terminal carbohydrate recognition domain.
Glycobiology, 19, 2009
|
|
2Z5I
 
 | | Crystal structure of the head-to-tail junction of tropomyosin | | Descriptor: | General control protein GCN4 and Tropomyosin alpha-1 chain, MAGNESIUM ION, Tropomyosin alpha-1 chain and General control protein GCN4 | | Authors: | Murakami, K, Nozawa, K, Tomii, K, Kudou, N, Igarashi, N, Shirakihara, Y, Wakatsuki, S, Stewart, M, Yasunaga, T, Wakabayashi, T. | | Deposit date: | 2007-07-12 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for tropomyosin overlap in thin (actin) filaments and the generation of a molecular swivel by troponin-T
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2ZRT
 
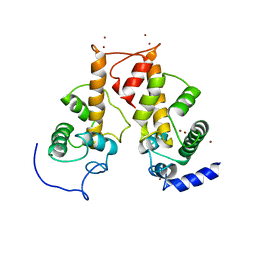 | | Crystal structure of Zn2+-bound form of des3-23ALG-2 | | Descriptor: | Programmed cell death protein 6, ZINC ION | | Authors: | Suzuki, H, Kawasaki, M, Kakiuchi, T, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystallization and X-ray diffraction analysis of N-terminally truncated human ALG-2
ACTA CRYSTALLOGR.,SECT.F, 64, 2008
|
|
2ZRS
 
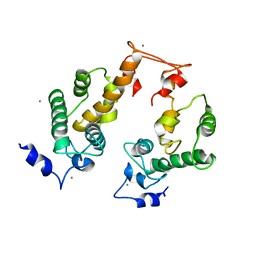 | | Crystal structure of Ca2+-bound form of des3-23ALG-2 | | Descriptor: | CALCIUM ION, Programmed cell death protein 6 | | Authors: | Suzuki, H, Kawasaki, M, Kakiuchi, T, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystallization and X-ray diffraction analysis of N-terminally truncated human ALG-2
ACTA CRYSTALLOGR.,SECT.F, 64, 2008
|
|
2ZHM
 
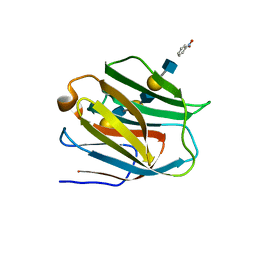 | | Crystal structure of human galectin-9 N-terminal CRD in complex with N-acetyllactosamine trimer (crystal 1) | | Descriptor: | (R)-1-PARA-NITRO-PHENYL-2-AZIDO-ETHANOL, Galectin-9, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nagae, M, Nishi, N, Murata, T, Usui, T, Nakamura, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2008-02-06 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural analysis of the recognition mechanism of poly-N-acetyllactosamine by the human galectin-9 N-terminal carbohydrate recognition domain.
Glycobiology, 19, 2009
|
|
2ZNE
 
 | | Crystal structure of Zn2+-bound form of des3-23ALG-2 complexed with Alix ABS peptide | | Descriptor: | 16-meric peptide from Programmed cell death 6-interacting protein, Programmed cell death protein 6, SODIUM ION, ... | | Authors: | Suzuki, H, Kawasaki, M, Inuzuka, T, Kakiuchi, T, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Ca(2+)-Dependent Formation of ALG-2/Alix Peptide Complex: Ca(2+)/EF3-Driven Arginine Switch Mechanism
Structure, 16, 2008
|
|
2ZHN
 
 | | Crystal structure of human galectin-9 N-terminal CRD in complex with N-acetyllactosamine trimer (crystal 2) | | Descriptor: | (R)-1-PARA-NITRO-PHENYL-2-AZIDO-ETHANOL, GALECTIN-9, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Nagae, M, Nishi, N, Murata, T, Usui, T, Nakamura, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2008-02-06 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural analysis of the recognition mechanism of poly-N-acetyllactosamine by the human galectin-9 N-terminal carbohydrate recognition domain.
Glycobiology, 19, 2009
|
|
2ZN8
 
 | | Crystal structure of Zn2+-bound form of ALG-2 | | Descriptor: | Programmed cell death protein 6, SODIUM ION, ZINC ION | | Authors: | Suzuki, H, Kawasaki, M, Inuzuka, T, Kakiuchi, T, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Ca(2+)-Dependent Formation of ALG-2/Alix Peptide Complex: Ca(2+)/EF3-Driven Arginine Switch Mechanism
Structure, 16, 2008
|
|
2ZND
 
 | | Crystal structure of Ca2+-free form of des3-20ALG-2 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, PHOSPHATE ION, Programmed cell death protein 6, ... | | Authors: | Suzuki, H, Kawasaki, M, Inuzuka, T, Kakiuchi, T, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Ca(2+)-Dependent Formation of ALG-2/Alix Peptide Complex: Ca(2+)/EF3-Driven Arginine Switch Mechanism
Structure, 16, 2008
|
|
2Z9Y
 
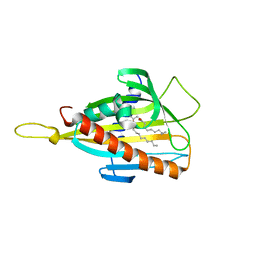 | | Crystal structure of CERT START domain in complex with C10-diacylglycerol | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl didecanoate, Lipid-transfer protein CERT | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2007-09-26 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2ZAN
 
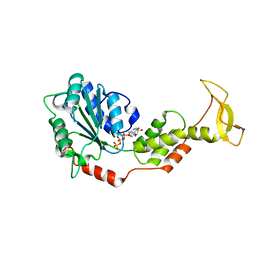 | | Crystal structure of mouse SKD1/VPS4B ATP-form | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Vacuolar protein sorting-associating protein 4B | | Authors: | Inoue, M, Kawasaki, M, Kamikubo, H, Kataoka, M, Kato, R, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2007-10-08 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Nucleotide-dependent conformational changes and assembly of the AAA ATPase SKD1/VPS4B
Traffic, 9, 2008
|
|
3AAK
 
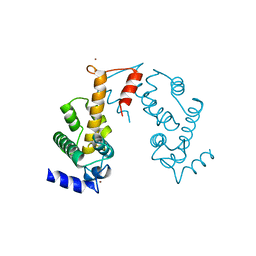 | | Crystal structure of Zn2+-bound form of des3-20ALG-2F122A | | Descriptor: | Programmed cell death protein 6, ZINC ION | | Authors: | Inuzuka, T, Suzuki, H, Kawasaki, M, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2009-11-19 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for defect in Alix-binding by alternatively spliced isoform of ALG-2 (ALG-2DeltaGF122) and structural roles of F122 in target recognition
Bmc Struct.Biol., 10, 2010
|
|
3AAJ
 
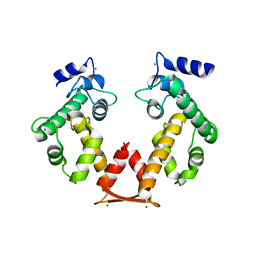 | | Crystal structure of Ca2+-bound form of des3-23ALG-2deltaGF122 | | Descriptor: | CALCIUM ION, Programmed cell death protein 6 | | Authors: | Suzuki, H, Inuzuka, T, Kawasaki, M, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2009-11-19 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for defect in Alix-binding by alternatively spliced isoform of ALG-2 (ALG-2DeltaGF122) and structural roles of F122 in target recognition
Bmc Struct.Biol., 10, 2010
|
|
