3ZHT
 
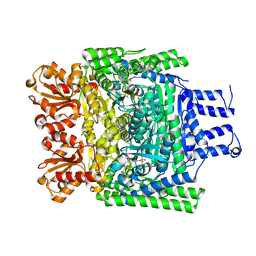 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD, first post-decarboxylation intermediate from 2-oxoadipate | | Descriptor: | (5S)-5-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-4-methyl-5-(2-{[(phosphonatooxy)phosphinato]oxy}ethyl)-1,3-thiazol-3-ium-2-yl}-5-hydroxypentanoate, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Barilone, N, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2012-12-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Dual Conformation of the Post-Decarboxylation Intermediate is Associated with Distinct Enzyme States in Mycobacterial Alpha-Ketoglutarate Decarboxylase (Kgd).
Biochem.J., 457, 2014
|
|
3ZHQ
 
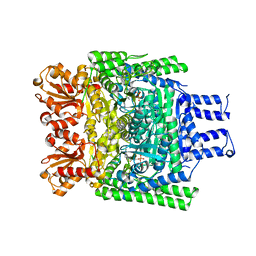 | | Crystal structure of the H747A mutant of the SucA domain of Mycobacterium smegmatis KGD | | Descriptor: | CALCIUM ION, MAGNESIUM ION, MULTIFUNCTIONAL 2-OXOGLUTARATE METABOLISM ENZYME, ... | | Authors: | Wagner, T, Barilone, N, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2012-12-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Dual Conformation of the Post-Decarboxylation Intermediate is Associated with Distinct Enzyme States in Mycobacterial Alpha-Ketoglutarate Decarboxylase (Kgd).
Biochem.J., 457, 2014
|
|
3ZHV
 
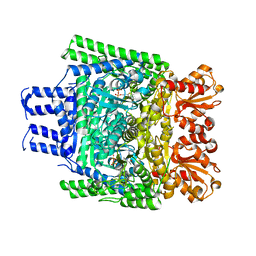 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD, post-decarboxylation intermediate from pyruvate (2-hydroxyethyl-ThDP) | | Descriptor: | 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-2-[(1S)-1-oxidanylethyl]-1,3-thiazol-3-ium-5-yl]ethyl phosphono hydrogen phosphate, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Barilone, N, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2012-12-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Dual Conformation of the Post-Decarboxylation Intermediate is Associated with Distinct Enzyme States in Mycobacterial Alpha-Ketoglutarate Decarboxylase (Kgd).
Biochem.J., 457, 2014
|
|
5M06
 
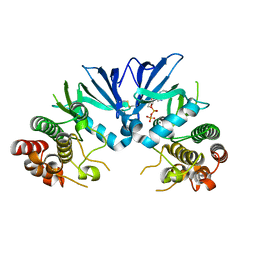 | | Crystal structure of Mycobacterium tuberculosis PknI kinase domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Serine/threonine-protein kinase PknI | | Authors: | Wagner, T, Lisa, M.N, Alexandre, M, Barilone, N, Raynal, B, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2016-10-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of PknI from Mycobacterium tuberculosis shows an inactive, pseudokinase-like conformation.
FEBS J., 284, 2017
|
|
3ZHU
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD, second post-decarboxylation intermediate from 2-oxoadipate | | Descriptor: | (5R)-5-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-4-methyl-5-(2-{[(phosphonatooxy)phosphinato]oxy}ethyl)-1,3-thiazol-3-ium-2-yl}-5-hydroxypentanoate, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Barilone, N, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2012-12-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Dual Conformation of the Post-Decarboxylation Intermediate is Associated with Distinct Enzyme States in Mycobacterial Alpha-Ketoglutarate Decarboxylase (Kgd).
Biochem.J., 457, 2014
|
|
3ZHS
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD, first post-decarboxylation intermediate from alpha-ketoglutarate | | Descriptor: | (4S)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3lambda~5~-thiazol-2-yl}-4-hydroxybutanoic acid, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Barilone, N, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2012-12-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Dual Conformation of the Post-Decarboxylation Intermediate is Associated with Distinct Enzyme States in Mycobacterial Alpha-Ketoglutarate Decarboxylase (Kgd).
Biochem.J., 457, 2014
|
|
6EP8
 
 | | InhA Y158F mutant in complex with NADH from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Enoyl-[acyl-carrier-protein] reductase [NADH], GLYCEROL, ... | | Authors: | Wagner, T, Voegeli, B, Rosenthal, R.G, Stoffel, G, Shima, S, Kiefer, P, Cortina, N, Erb, T.J. | | Deposit date: | 2017-10-11 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | InhA, the enoyl-thioester reductase fromMycobacterium tuberculosisforms a covalent adduct during catalysis.
J. Biol. Chem., 293, 2018
|
|
3ZHR
 
 | | Crystal structure of the H747A mutant of the SucA domain of Mycobacterium smegmatis KGD showing the active site lid closed | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Barilone, N, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2012-12-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Dual Conformation of the Post-Decarboxylation Intermediate is Associated with Distinct Enzyme States in Mycobacterial Alpha-Ketoglutarate Decarboxylase (Kgd).
Biochem.J., 457, 2014
|
|
6S6Y
 
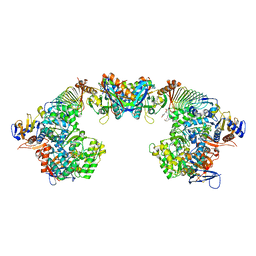 | | X-ray crystal structure of the formyltransferase/hydrolase complex (FhcABCD) from Methylorubrum extorquens in complex with methylofuran | | Descriptor: | (2~{S})-3-[4-[[5-(aminomethyl)furan-3-yl]methoxy]phenyl]-2-(methylamino)propanoic acid, 1,2-ETHANEDIOL, AMINO GROUP, ... | | Authors: | Wagner, T, Hemmann, J.L, Shima, S, Vorholt, J. | | Deposit date: | 2019-07-04 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Methylofuran is a prosthetic group of the formyltransferase/hydrolase complex and shuttles one-carbon units between two active sites.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6TLK
 
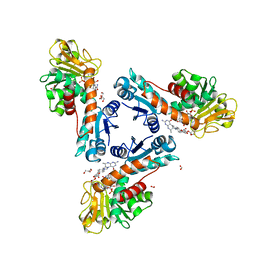 | | Structure of methylene-tetrahydromethanopterin dehydrogenase from Methylorubrum extorquens AM1 in an open conformation containing NADP+ and methylene-H4MPT | | Descriptor: | 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, Bifunctional protein MdtA, CHLORIDE ION, ... | | Authors: | Wagner, T, Huang, G, Demmer, U, Warkentin, E, Ermler, U, Shima, S. | | Deposit date: | 2019-12-02 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Hydride Transfer Process in NADP-dependent Methylene-tetrahydromethanopterin Dehydrogenase.
J.Mol.Biol., 432, 2020
|
|
6GGU
 
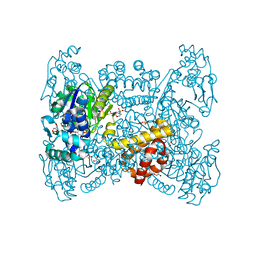 | | Crystal structure of native FE-hydrogenase from Methanothermobacter marburgensis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5,10-methenyltetrahydromethanopterin hydrogenase, GLYCEROL, ... | | Authors: | Wagner, T, Huang, G, Ermler, U, Shima, S. | | Deposit date: | 2018-05-03 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Self-protection of the catalytic iron center of a methanogenic [Fe]-hydrogenase via a dynamic dimer-to-hexamer transformation
To Be Published
|
|
6YTT
 
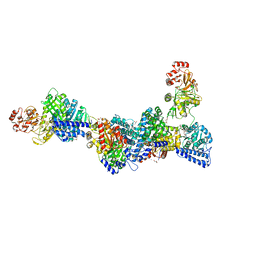 | | CO-dehydrogenase/Acetyl-CoA synthase (CODH/ACS) from Clostridium autoethanogenum at 3.0-A resolution | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CO dehydrogenase/acetyl-CoA synthase complex, beta subunit, ... | | Authors: | Wagner, T, Lemaire, O.N. | | Deposit date: | 2020-04-24 | | Release date: | 2020-11-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Gas channel rerouting in a primordial enzyme: Structural insights of the carbon-monoxide dehydrogenase/acetyl-CoA synthase complex from the acetogen Clostridium autoethanogenum.
Biochim Biophys Acta Bioenerg, 1862, 2020
|
|
1GG0
 
 | | CRYSTAL STRUCTURE ANALYSIS OF KDOP SYNTHASE AT 3.0 A | | Descriptor: | 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE SYNTHASE, PHOSPHATE ION | | Authors: | Wagner, T, Kretsinger, R.H, Bauerle, R, Tolbert, W.D. | | Deposit date: | 2000-08-04 | | Release date: | 2000-10-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 3-Deoxy-D-manno-octulosonate-8-phosphate synthase from Escherichia coli. Model of binding of phosphoenolpyruvate and D-arabinose-5-phosphate.
J.Mol.Biol., 301, 2000
|
|
5LB9
 
 | | Structure of the T175V Etr1p mutant in the monoclinic form P21 | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH, B-specific] 1, mitochondrial, ... | | Authors: | Wagner, T, Rosenthal, R.G, Voegeli, B, Shima, S, Erb, T.J. | | Deposit date: | 2016-06-15 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A conserved threonine prevents self-intoxication of enoyl-thioester reductases.
Nat. Chem. Biol., 13, 2017
|
|
5LBX
 
 | | Structure of the T175V Etr1p mutant in the trigonal form P312 in complex with NADP and crotonyl-CoA | | Descriptor: | CHLORIDE ION, CROTONYL COENZYME A, Enoyl-[acyl-carrier-protein] reductase [NADPH, ... | | Authors: | Wagner, T, Rosenthal, R.G, Voegeli, B, Shima, S, Erb, T.J. | | Deposit date: | 2016-06-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A conserved threonine prevents self-intoxication of enoyl-thioester reductases.
Nat. Chem. Biol., 13, 2017
|
|
5N1Q
 
 | | METHYL-COENZYME M REDUCTASE III FROM METHANOTHERMOCOCCUS THERMOLITHOTROPHICUS AT 1.9 A RESOLUTION | | Descriptor: | 1-THIOETHANESULFONIC ACID, Coenzyme B, FACTOR 430, ... | | Authors: | Wagner, T, Wegner, C.E, Ermler, U, Shima, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phylogenetic and Structural Comparisons of the Three Types of Methyl Coenzyme M Reductase from Methanococcales and Methanobacteriales.
J.Bacteriol., 199, 2017
|
|
7B2C
 
 | | Crystal structure of the ethyl-coenzyme M reductase from Candidatus Ethanoperedens thermophilum gassed with xenon | | Descriptor: | (2R)-2-[(2S)-2-[(2S)-2-oxidanylpropoxy]propoxy]propan-1-ol, 1-THIOETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Wagner, T, Lemaire, O.N, Engilberge, S. | | Deposit date: | 2020-11-26 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a key enzyme for anaerobic ethane activation.
Science, 373, 2021
|
|
5O4N
 
 | | Apo HcgC from Methanococcus maripaludis soaked with SAH and pyridinol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-carboxy methyl-4-hydroxy-2-pyridinol, DIMETHYL SULFOXIDE, ... | | Authors: | Wagner, T, Bai, L, Xu, T, Hu, X, Ermler, U, Shima, S. | | Deposit date: | 2017-05-29 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Water-Bridged H-Bonding Network Contributes to the Catalysis of the SAM-Dependent C-Methyltransferase HcgC.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5A8R
 
 | |
5A0Y
 
 | |
5ODQ
 
 | | Heterodisulfide reductase / [NiFe]-hydrogenase complex from Methanothermococcus thermolithotrophicus soaked with bromoethanesulfonate. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-bromanylethanesulfonic acid, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | Wagner, T, Koch, J, Ermler, U, Shima, S. | | Deposit date: | 2017-07-06 | | Release date: | 2017-08-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction.
Science, 357, 2017
|
|
5ODC
 
 | | Heterodisulfide reductase / [NiFe]-hydrogenase complex from Methanothermococcus thermolithotrophicus at 2.3 A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETATE ION, ... | | Authors: | Wagner, T, Koch, J, Ermler, U, Shima, S. | | Deposit date: | 2017-07-05 | | Release date: | 2017-08-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction.
Science, 357, 2017
|
|
5ODI
 
 | | Heterodisulfide reductase / [NiFe]-hydrogenase complex from Methanothermococcus thermolithotrophicus cocrystallized with CoM-SH | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | Wagner, T, Koch, J, Ermler, U, Shima, S. | | Deposit date: | 2017-07-05 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction.
Science, 357, 2017
|
|
5ODR
 
 | | Heterodisulfide reductase / [NiFe]-hydrogenase complex from Methanothermococcus thermolithotrophicus soaked with heterodisulfide for 2 minutes. | | Descriptor: | 1-THIOETHANESULFONIC ACID, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETATE ION, ... | | Authors: | Wagner, T, Koch, J, Ermler, U, Shima, S. | | Deposit date: | 2017-07-06 | | Release date: | 2017-08-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction.
Science, 357, 2017
|
|
6YUA
 
 | |
