4ZO2
 
 | | AidC, a Dizinc Quorum-Quenching Lactonase | | Descriptor: | Acylhomoserine lactonase, ZINC ION | | Authors: | Mascarenhas, R, Thomas, P.W, Wu, C.-X, Nocek, B.P, Hoang, Q, Fast, W, Liu, D. | | Deposit date: | 2015-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Structural and Biochemical Characterization of AidC, a Quorum-Quenching Lactonase with Atypical Selectivity.
Biochemistry, 54, 2015
|
|
1X9X
 
 | | Solution Structure of Dimeric SAM Domain from MAPKKK Ste11 | | Descriptor: | Serine/threonine-protein kinase STE11 | | Authors: | Bhattacharjya, S, Xu, P, Gingras, R, Shaykhutdinov, R, Wu, C, Whiteway, M, Ni, F. | | Deposit date: | 2004-08-24 | | Release date: | 2005-08-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the dimeric SAM domain of MAPKKK Ste11 and its interactions with the adaptor protein Ste50 from the budding yeast: implications for Ste11 activation and signal transmission through the Ste50-Ste11 complex.
J.Mol.Biol., 344, 2004
|
|
1NYP
 
 | | 4th LIM domain of PINCH protein | | Descriptor: | PINCH protein, ZINC ION | | Authors: | Velyvis, A, Vaynberg, J, Vinogradova, O, Zhang, Y, Wu, C, Qin, J. | | Deposit date: | 2003-02-13 | | Release date: | 2003-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and functional insights into PINCH LIM4 domain-mediated integrin signaling
Nat.Struct.Biol., 10, 2003
|
|
2VXU
 
 | | Crystal structure of murine reference antibody 125-2H Fab fragment | | Descriptor: | MURINE IGG 125-2H | | Authors: | Argiriadi, M.A, Xiang, T, Wu, C, Ghayur, T, Borhani, D.W. | | Deposit date: | 2008-07-10 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Unusual Water-Mediated Antigenic Recognition of the Proinflammatory Cytokine Interleukin-18.
J.Biol.Chem., 284, 2009
|
|
5CHL
 
 | | Structural basis of H2A.Z recognition by YL1 histone chaperone component of SRCAP/SWR1 chromatin remodeling complex | | Descriptor: | Histone H2A.Z, Vacuolar protein sorting-associated protein 72 homolog | | Authors: | Shan, S, Liang, X, Pan, L, Wu, C, Zhou, Z. | | Deposit date: | 2015-07-10 | | Release date: | 2016-03-09 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Structural basis of H2A.Z recognition by SRCAP chromatin-remodeling subunit YL1
Nat.Struct.Mol.Biol., 23, 2016
|
|
2PO7
 
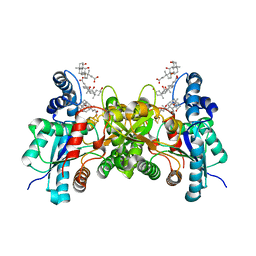 | | Crystal structure of human ferrochelatase mutant with His 341 replaced by Cys | | Descriptor: | CHOLIC ACID, FE2/S2 (INORGANIC) CLUSTER, Ferrochelatase, ... | | Authors: | Dailey, H.A, Wu, C.-K, Horanyi, P, Medlock, A.E, Najahi-Missaoui, W, Burden, A, Dailey, T.A, Rose, J.P. | | Deposit date: | 2007-04-25 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Altered orientation of active site residues in variants of human ferrochelatase. Evidence for a hydrogen bond network involved in catalysis.
Biochemistry, 46, 2007
|
|
2L5A
 
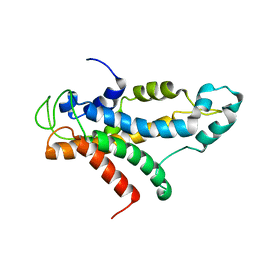 | | Structural basis for recognition of centromere specific histone H3 variant by nonhistone Scm3 | | Descriptor: | Histone H3-like centromeric protein CSE4, Protein SCM3, Histone H4 | | Authors: | Zhou, Z, Feng, H, Zhou, B, Ghirlando, R, Hu, K, Zwolak, A, Jenkins, L, Xiao, H, Tjandra, N, Wu, C, Bai, Y. | | Deposit date: | 2010-10-28 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3.
Nature, 472, 2011
|
|
3L86
 
 | |
7R81
 
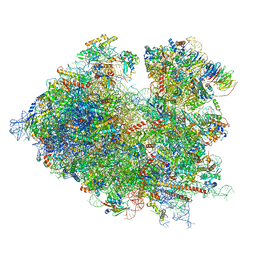 | | Structure of the translating Neurospora crassa ribosome arrested by cycloheximide | | Descriptor: | 18S rRNA, 26S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Shen, L, Su, Z, Yang, K, Wu, C, Becker, T, Bell-Pedersen, D, Zhang, J, Sachs, M.S. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of the translating Neurospora ribosome arrested by cycloheximide
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4ZO3
 
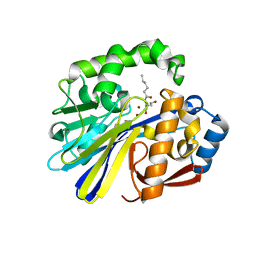 | | AidC, a Dizinc Quorum-Quenching Lactonase, in complex with a product N-hexnoyl-L-homoserine | | Descriptor: | Acylhomoserine lactonase, N-hexanoyl-L-homoserine, ZINC ION | | Authors: | Mascarenhas, R, Thomas, P.W, Wu, C.-X, Nocek, B.P, Hoang, Q, Fast, W, Liu, D. | | Deposit date: | 2015-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural and Biochemical Characterization of AidC, a Quorum-Quenching Lactonase with Atypical Selectivity.
Biochemistry, 54, 2015
|
|
2JSS
 
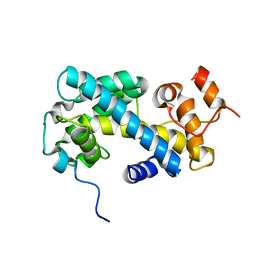 | | NMR structure of chaperone Chz1 complexed with histone H2A.Z-H2B | | Descriptor: | Chimera of Histone H2B.1 and Histone H2A.Z, Uncharacterized protein YER030W | | Authors: | Zhou, Z, Feng, H, Hansen, D.F, Kato, H, Luk, E, Freedberg, D.I, Kay, L.E, Wu, C, Bai, Y. | | Deposit date: | 2007-07-11 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of chaperone Chz1 complexed with histones H2A.Z-H2B.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2LAS
 
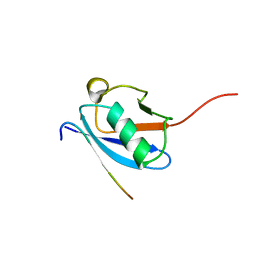 | | Molecular Determinants of Paralogue-Specific SUMO-SIM Recognition | | Descriptor: | M-IR2_peptide, Small ubiquitin-related modifier 1 | | Authors: | Namanja, A, Li, Y, Su, Y, Wong, S, Lu, J, Colson, L, Wu, C, Li, S, Chen, Y. | | Deposit date: | 2011-03-20 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into High Affinity Small Ubiquitin-like Modifier (SUMO) Recognition by SUMO-interacting Motifs (SIMs) Revealed by a Combination of NMR and Peptide Array Analysis.
J.Biol.Chem., 287, 2012
|
|
7W8O
 
 | |
7W8K
 
 | |
7W8R
 
 | |
7W96
 
 | |
7WE3
 
 | |
7WEI
 
 | |
6YPZ
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
6YQ6
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
6YQ3
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | (3~{R})-8-methoxy-3-methyl-3,6-bis(oxidanyl)-2,4-dihydrobenzo[a]anthracene-1,7,12-trione, 1,2-ETHANEDIOL, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
6YQ0
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | (3~{R})-8-methoxy-3-methyl-3-oxidanyl-2,4-dihydrobenzo[a]anthracene-1,7,12-trione, 1,2-ETHANEDIOL, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
8BV0
 
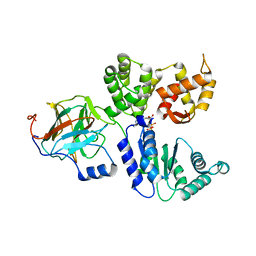 | | Binary complex between the NB-ARC domain from the Tomato immune receptor NRC1 and the SPRY domain-containing effector SS15 from the potato cyst nematode | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NRC1, Truncated secreted SPRY domain-containing protein 15 (Fragment) | | Authors: | Contreras, M.P, Pai, H, Muniyandi, S, Toghani, A, Lawson, D.M, Tumtas, Y, Duggan, C, Yuen, E.L.H, Stevenson, C.E.M, Harant, A, Wu, C.H, Bozkurt, T.O, Kamoun, S, Derevnina, L. | | Deposit date: | 2022-12-01 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Resurrection of plant disease resistance proteins via helper NLR bioengineering.
Sci Adv, 9, 2023
|
|
7W8Z
 
 | |
7W8T
 
 | |
