7Q6L
 
 | |
7QA2
 
 | |
3CP1
 
 | |
3LQJ
 
 | |
3LQI
 
 | |
3CYO
 
 | |
1OVL
 
 | | Crystal Structure of Nurr1 LBD | | Descriptor: | BROMIDE ION, IODIDE ION, Orphan nuclear receptor NURR1 (MSE 414, ... | | Authors: | Wang, Z, Liu, J, Walker, N. | | Deposit date: | 2003-03-26 | | Release date: | 2003-06-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Function of Nurr1 identifies a Class of Ligand-Independent Nuclear Receptors
Nature, 423, 2003
|
|
1PBP
 
 | | FINE TUNING OF THE SPECIFICITY OF THE PERIPLASMIC PHOSPHATE TRANSPORT RECEPTOR: SITE-DIRECTED MUTAGENESIS, LIGAND BINDING, AND CRYSTALLOGRAPHIC STUDIES | | Descriptor: | PHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Wang, Z, Choudhary, A, Ledvina, P.S, Quiocho, F.A. | | Deposit date: | 1994-07-20 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fine tuning the specificity of the periplasmic phosphate transport receptor. Site-directed mutagenesis, ligand binding, and crystallographic studies.
J.Biol.Chem., 269, 1994
|
|
3NP2
 
 | | Crystal Structure of Pd(allyl)/apo-E45C/C48A-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Wang, Z, Ueno, T, Abe, S, Takezawa, Y, Aoyagi, H, Hikage, T, Watanabe, Y, Kitagawa, S. | | Deposit date: | 2010-06-27 | | Release date: | 2010-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Definite coordination arrangement of organometallic palladium complexes accumulated on the designed interior surface of apo-ferritin.
Chem.Commun.(Camb.), 47, 2011
|
|
3V3L
 
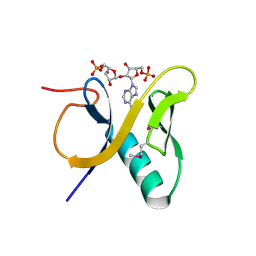 | | Crystal structure of human RNF146 WWE domain in complex with iso-ADPRibose | | Descriptor: | 2'-O-(5-O-phosphono-alpha-D-ribofuranosyl)adenosine 5'-(dihydrogen phosphate), E3 ubiquitin-protein ligase RNF146 | | Authors: | Wang, Z, Cheng, Z, Xu, W. | | Deposit date: | 2011-12-13 | | Release date: | 2012-02-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination.
Genes Dev., 26, 2012
|
|
5CL1
 
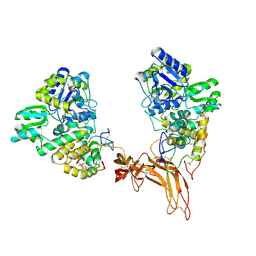 | | Complex structure of Norrin with human Frizzled 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Frizzled-4, Maltose-binding periplasmic protein,Norrin | | Authors: | Wang, Z, Ke, J, Shen, G, Cheng, Z, Xu, H.E, Xu, W. | | Deposit date: | 2015-07-16 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of the Norrin-Frizzled 4 interaction.
Cell Res., 25, 2015
|
|
3NP0
 
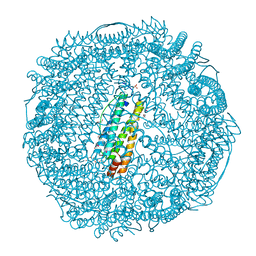 | | Crystal Structure of Pd(allyl)/apo-E45C/H49A/R52H-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Wang, Z, Ueno, T, Abe, S, Takezawa, Y, Aoyagi, H, Hikage, T, Watanabe, Y, Kitagawa, S. | | Deposit date: | 2010-06-27 | | Release date: | 2010-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Definite coordination arrangement of organometallic palladium complexes accumulated on the designed interior surface of apo-ferritin.
Chem.Commun.(Camb.), 47, 2011
|
|
3NOZ
 
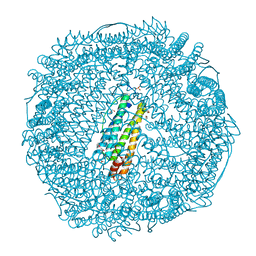 | | Crystal Structure of Pd(allyl)/apo-E45C/R52H-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Wang, Z, Ueno, T, Abe, S, Takezawa, Y, Aoyagi, H, Hikage, T, Watanabe, Y, Kitagawa, S. | | Deposit date: | 2010-06-27 | | Release date: | 2010-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Definite coordination arrangement of organometallic palladium complexes accumulated on the designed interior surface of apo-ferritin.
Chem.Commun.(Camb.), 47, 2011
|
|
4ERK
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE ERK2/OLOMOUCINE | | Descriptor: | EXTRACELLULAR REGULATED KINASE 2, OLOMOUCINE, SULFATE ION | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-09 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
3ERK
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE ERK2/SB220025 | | Descriptor: | 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, EXTRACELLULAR REGULATED KINASE 2 | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-09 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
6LK9
 
 | | Coho salmon ferritin | | Descriptor: | Coho salmon ferritin, FE (III) ION | | Authors: | Wang, Z, Zang, J, Li, H, Tan, X, Du, M. | | Deposit date: | 2019-12-18 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Coho salmon ferritin
To Be Published
|
|
1ZJ6
 
 | | Crystal structure of human ARL5 | | Descriptor: | ADP-ribosylation factor-like protein 5, GUANOSINE-3'-MONOPHOSPHATE-5'-DIPHOSPHATE, SULFATE ION | | Authors: | Wang, Z.X, Shi, L, Liu, J.F, An, X.M, Chang, W.R, Liang, D.C. | | Deposit date: | 2005-04-28 | | Release date: | 2005-06-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0A crystal structure of human ARL5-GDP3'P, a novel member of the small GTP-binding proteins
Biochem.Biophys.Res.Commun., 332, 2005
|
|
2DW3
 
 | |
3GZ9
 
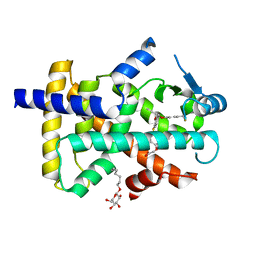 | | Crystal Structure of Peroxisome Proliferator-Activated Receptor Delta (PPARd) in Complex with a Full Agonist | | Descriptor: | (2,3-dimethyl-4-{[2-(prop-2-yn-1-yloxy)-4-{[4-(trifluoromethyl)phenoxy]methyl}phenyl]sulfanyl}phenoxy)acetic acid, Peroxisome proliferator-activated receptor delta, heptyl beta-D-glucopyranoside | | Authors: | Wang, Z, Sudom, A, Walker, N.P. | | Deposit date: | 2009-04-06 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a PPARdelta agonist with partial agonistic activity on PPARgamma.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3H0A
 
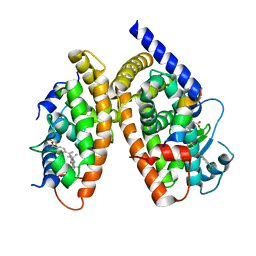 | | Crystal Structure of Peroxisome Proliferator-Activated Receptor Gamma (PPARg) and Retinoic Acid Receptor Alpha (RXRa) in Complex with 9-cis Retinoic Acid, Co-activator Peptide, and a Partial Agonist | | Descriptor: | 4-[1-(3,5,5,8,8-pentamethyl-5,6,7,8-tetrahydronaphthalen-2-yl)ethenyl]benzoic acid, Nuclear receptor coactivator 1, Co-activator Peptide, ... | | Authors: | Wang, Z, Sudom, A, Walker, N.P. | | Deposit date: | 2009-04-08 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of a PPARdelta agonist with partial agonistic activity on PPARgamma.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3J7N
 
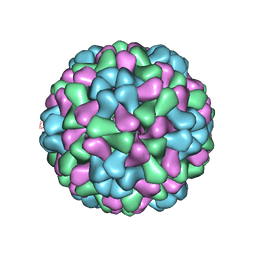 | | Virus model of brome mosaic virus (second half data set) | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
3J7L
 
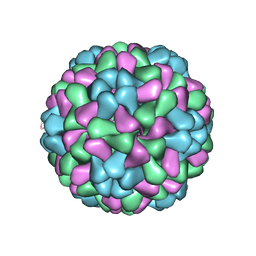 | | Full virus map of brome mosaic virus | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
7EW5
 
 | |
3J7M
 
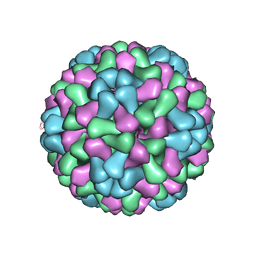 | | Virus model of brome mosaic virus (first half data set) | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
3K8S
 
 | | Crystal Structure of PPARg in complex with T2384 | | Descriptor: | 2-chloro-N-{3-chloro-4-[(5-chloro-1,3-benzothiazol-2-yl)sulfanyl]phenyl}-4-(trifluoromethyl)benzenesulfonamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Wang, Z. | | Deposit date: | 2009-10-14 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | T2384, a novel antidiabetic agent with unique peroxisome proliferator-activated receptor gamma binding properties
J.Biol.Chem., 283, 2008
|
|
