5DQT
 
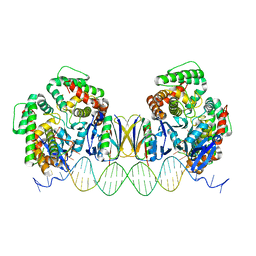 | | Crystal Structure of Cas-DNA-22 complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (33-MER), ... | | Authors: | Wang, J, Li, J, Zhao, H, Sheng, G, Wang, M, Yin, M, Wang, Y. | | Deposit date: | 2015-09-15 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems.
Cell, 163, 2015
|
|
7R76
 
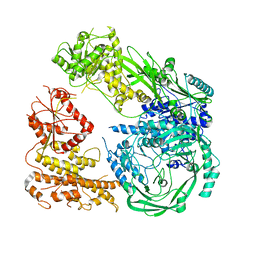 | | Cryo-EM structure of DNMT5 in apo state | | Descriptor: | DNA repair protein Rad8, ZINC ION | | Authors: | Wang, J, Patel, D.J. | | Deposit date: | 2021-06-24 | | Release date: | 2022-02-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into DNMT5-mediated ATP-dependent high-fidelity epigenome maintenance.
Mol.Cell, 82, 2022
|
|
7R77
 
 | |
7R78
 
 | |
3QZQ
 
 | | Human enterovirus 71 3C protease mutant E71D in complex with rupintrivir | | Descriptor: | 3C protein, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Wang, J, Fan, T, Yao, X, Wu, Z, Guo, L, Lei, X, Wang, J, Wang, M, Jin, Q, Cui, S. | | Deposit date: | 2011-03-07 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7001 Å) | | Cite: | Crystal Structures of Enterovirus 71 3C Protease Complexed with Rupintrivir Reveal the Roles of Catalytically Important Residues.
J.Virol., 85, 2011
|
|
3R0F
 
 | | Human enterovirus 71 3C protease mutant H133G in complex with rupintrivir | | Descriptor: | 1,2-ETHANEDIOL, 3C protein, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Wang, J, Fan, T, Yao, X, Wu, Z, Guo, L, Lei, X, Wang, J, Wang, M, Jin, Q, Cui, S. | | Deposit date: | 2011-03-08 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3083 Å) | | Cite: | Crystal Structures of Enterovirus 71 3C Protease Complexed with Rupintrivir Reveal the Roles of Catalytically Important Residues.
J.Virol., 85, 2011
|
|
3QZR
 
 | | Human enterovirus 71 3C protease mutant E71A in complex with rupintrivir | | Descriptor: | 1,2-ETHANEDIOL, 3C protein, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Wang, J, Fan, T, Yao, X, Wu, Z, Guo, L, Lei, X, Wang, J, Wang, M, Jin, Q, Cui, S. | | Deposit date: | 2011-03-07 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.039 Å) | | Cite: | Crystal Structures of Enterovirus 71 3C Protease Complexed with Rupintrivir Reveal the Roles of Catalytically Important Residues.
J.Virol., 85, 2011
|
|
3OSY
 
 | |
1NFD
 
 | | AN ALPHA-BETA T CELL RECEPTOR (TCR) HETERODIMER IN COMPLEX WITH AN ANTI-TCR FAB FRAGMENT DERIVED FROM A MITOGENIC ANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H57 FAB, N15 ALPHA-BETA T-CELL RECEPTOR | | Authors: | Wang, J.-H, Lim, K, Smolyar, A, Teng, M.-K, Sacchittini, J, Reinherz, E.L. | | Deposit date: | 1997-08-04 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atomic structure of an alphabeta T cell receptor (TCR) heterodimer in complex with an anti-TCR fab fragment derived from a mitogenic antibody.
EMBO J., 17, 1998
|
|
1HQY
 
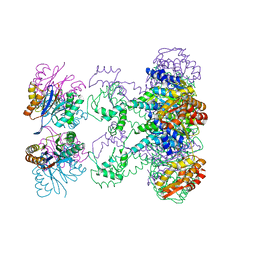 | | Nucleotide-Dependent Conformational Changes in a Protease-Associated ATPase HslU | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK LOCUS HSLU, HEAT SHOCK LOCUS HSLV | | Authors: | Wang, J, Song, J.J, Seong, I.S, Franklin, M.C, Kamtekar, S, Eom, S.H, Chung, C.H. | | Deposit date: | 2000-12-20 | | Release date: | 2001-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU.
Structure, 9, 2001
|
|
1HT1
 
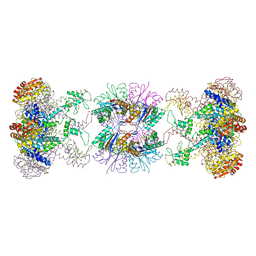 | | Nucleotide-Dependent Conformational Changes in a Protease-Associated ATPase HslU | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK LOCUS HSLU, HEAT SHOCK LOCUS HSLV | | Authors: | Wang, J, Song, J.J, Seong, I.S, Franklin, M.C, Kamtekar, S, Eom, S.H, Chung, C.H. | | Deposit date: | 2000-12-27 | | Release date: | 2001-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU.
Structure, 9, 2001
|
|
1HT2
 
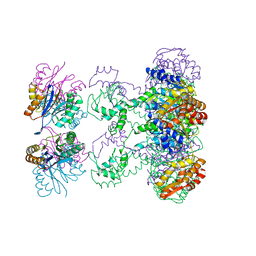 | | Nucleotide-Dependent Conformational Changes in a Protease-Associated ATPase HslU | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK LOCUS HSLU, HEAT SHOCK LOCUS HSLV | | Authors: | Wang, J, Song, J.J, Seong, I.S, Franklin, M.C, Kamtekar, S, Eom, S.H, Chung, C.H. | | Deposit date: | 2000-12-27 | | Release date: | 2001-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU.
Structure, 9, 2001
|
|
6LAF
 
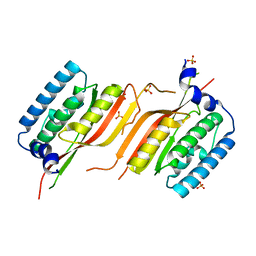 | | Crystal structure of the core domain of Amuc_1100 from Akkermansia muciniphila | | Descriptor: | Amuc_1100, SULFATE ION | | Authors: | Wang, J, Xiang, R, Zhang, M, Wang, M. | | Deposit date: | 2019-11-12 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | The variable oligomeric state of Amuc_1100 from Akkermansia muciniphila.
J.Struct.Biol., 212, 2020
|
|
6OM1
 
 | | Crystal structure of an atypical integrin | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, J.C, Springer, T.A. | | Deposit date: | 2019-04-17 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | General structural features that regulate integrin affinity revealed by atypical alpha V beta 8.
Nat Commun, 10, 2019
|
|
8KGE
 
 | |
8KHQ
 
 | | Bifunctional sulfoxide synthase OvoA_Th2 in complex with histidine and cysteine | | Descriptor: | 5-histidylcysteine sulfoxide synthase/putative 4-mercaptohistidine N1-methyltranferase, COBALT (II) ION, CYSTEINE, ... | | Authors: | Wang, J, Ye, K, Wang, X.Y, Yan, W.P. | | Deposit date: | 2023-08-22 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Biochemical and Structural Characterization of OvoA Th2 : A Mononuclear Nonheme Iron Enzyme from Hydrogenimonas thermophila for Ovothiol Biosynthesis.
Acs Catalysis, 13, 2023
|
|
4RP9
 
 | | Bacterial vitamin C transporter UlaA/SgaT in C2 form | | Descriptor: | ASCORBIC ACID, Ascorbate-specific permease IIC component UlaA, PENTAETHYLENE GLYCOL, ... | | Authors: | Wang, J.W. | | Deposit date: | 2014-10-29 | | Release date: | 2015-03-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Crystal structure of a phosphorylation-coupled vitamin C transporter.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4RP8
 
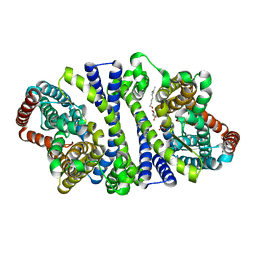 | | Bacterial vitamin C transporter UlaA/SgaT in P21 form | | Descriptor: | ASCORBIC ACID, Ascorbate-specific permease IIC component UlaA, nonyl beta-D-glucopyranoside | | Authors: | Wang, J.W. | | Deposit date: | 2014-10-29 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | Crystal structure of a phosphorylation-coupled vitamin C transporter.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6OM2
 
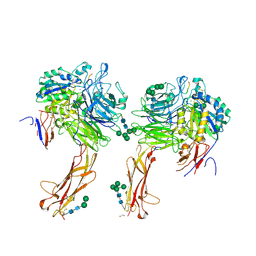 | |
2CCP
 
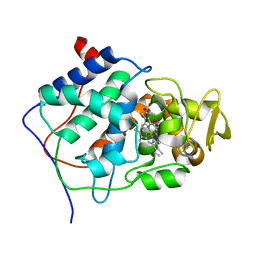 | | X-RAY STRUCTURES OF RECOMBINANT YEAST CYTOCHROME C PEROXIDASE AND THREE HEME-CLEFT MUTANTS PREPARED BY SITE-DIRECTED MUTAGENESIS | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, YEAST CYTOCHROME C PEROXIDASE | | Authors: | Wang, J, Mauro, J.M, Edwards, S.L, Oatley, S.J, Fishel, L.A, Ashford, V.A, Xuong, N.-H, Kraut, J. | | Deposit date: | 1990-02-28 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structures of recombinant yeast cytochrome c peroxidase and three heme-cleft mutants prepared by site-directed mutagenesis.
Biochemistry, 29, 1990
|
|
4Q5J
 
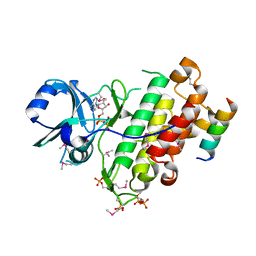 | | Crystal structure of SeMet derivative BRI1 in complex with BKI1 | | Descriptor: | BRI1 kinase inhibitor 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Protein BRASSINOSTEROID INSENSITIVE 1 | | Authors: | Wang, J, Wang, J, Chen, L, Wu, J.W, Wang, Z.X. | | Deposit date: | 2014-04-17 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.772 Å) | | Cite: | Structural insights into the negative regulation of BRI1 signaling by BRI1-interacting protein BKI1.
Cell Res., 24, 2014
|
|
4OH4
 
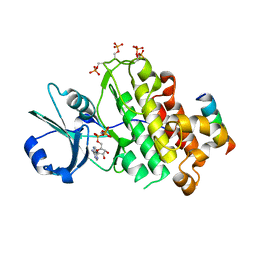 | | Crystal structure of BRI1 in complex with BKI1 | | Descriptor: | BRI1 kinase inhibitor 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Protein BRASSINOSTEROID INSENSITIVE 1 | | Authors: | Wang, J, Wang, J, Wu, J.W, Wang, Z.X. | | Deposit date: | 2014-01-17 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the negative regulation of BRI1 signaling by BRI1-interacting protein BKI1.
Cell Res., 24, 2014
|
|
8JVM
 
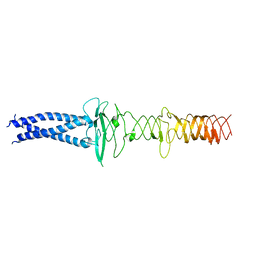 | | AHS-CSF domains of phage lambda tail | | Descriptor: | Tip attachment protein J | | Authors: | Wang, J. | | Deposit date: | 2023-06-28 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Architecture of the bacteriophage lambda tail.
Structure, 32, 2024
|
|
5GNF
 
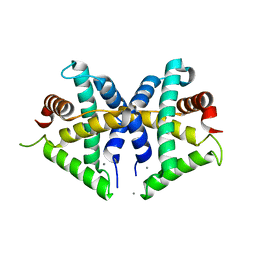 | | Crystal structure of anti-CRISPR protein AcrF3 | | Descriptor: | CALCIUM ION, Uncharacterized protein AcrF3 | | Authors: | Wang, J, Wang, Y. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A CRISPR evolutionary arms race: structural insights into viral anti-CRISPR/Cas responses
Cell Res., 26, 2016
|
|
6LYW
 
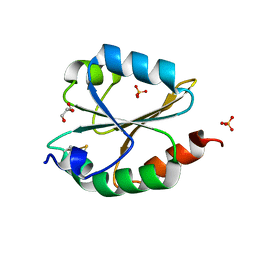 | | Structural insight into the biological functions of Arabidopsis thaliana ACHT1 | | Descriptor: | GLYCEROL, SULFATE ION, Thioredoxin-like 2-1, ... | | Authors: | Wang, J.C, Pan, W.M, Wang, M.Z, Zhang, M. | | Deposit date: | 2020-02-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into the biological functions of Arabidopsis thaliana ACHT1.
Int.J.Biol.Macromol., 158, 2020
|
|
