3W98
 
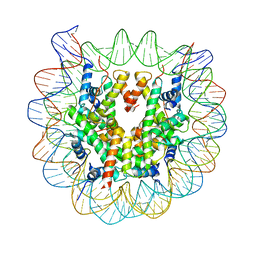 | | Crystal Structure of Human Nucleosome Core Particle lacking H3.1 N-terminal region | | Descriptor: | 146-mer DNA, Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Iwasaki, W, Miya, Y, Horikoshi, N, Osakabe, A, Tachiwana, H, Shibata, T, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2013-04-01 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Contribution of histone N-terminal tails to the structure and stability of nucleosomes
FEBS Open Bio, 3, 2013
|
|
8J97
 
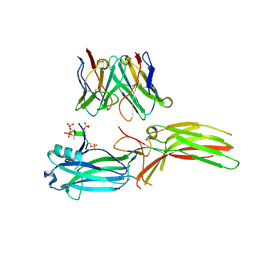 | | Structure of Muscarinic receptor (M2R) in complex with beta-arrestin1 (Local refine, cross-linked) | | Descriptor: | Beta-arrestin-1, Fab30 Heavy Chain, Fab30 Light Chain, ... | | Authors: | Maharana, J, Sano, F.K, Shihoya, W, Banerjee, R, Nureki, O, Shukla, A.K. | | Deposit date: | 2023-05-02 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
8JAF
 
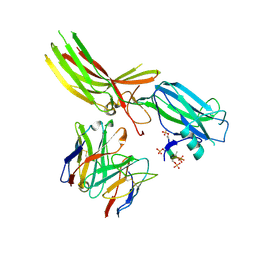 | | Structure of Muscarinic receptor (M2R) in complex with beta-arrestin1 (Local Refine, non-cross linked) | | Descriptor: | Beta-arrestin-1, Fab30 heavy chain, Fab30 light chain, ... | | Authors: | Maharana, J, Sano, F.K, Shihoya, W, Banerjee, R, Nureki, O, Shukla, A.K. | | Deposit date: | 2023-05-05 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
8J8R
 
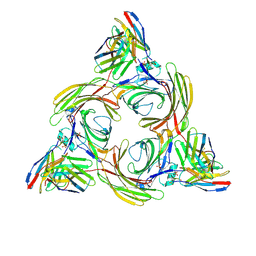 | | Structure of beta-arrestin2 in complex with M2Rpp | | Descriptor: | Beta-arrestin-2, Fab30 Heavy Chain, Fab30 Light Chain, ... | | Authors: | Maharana, J, Sano, F.K, Shihoya, W, Banerjee, R, Nureki, O, Shukla, A.K. | | Deposit date: | 2023-05-02 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
8JOU
 
 | | Fiber I and fiber-tail-adaptor of phage GP4 | | Descriptor: | Virion-associated phage protein, rope protein of phage GP4 | | Authors: | Liu, H, Chen, W. | | Deposit date: | 2023-06-08 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Asymmetric Structure of Podophage GP4 Reveals a Novel Architecture of Three Types of Tail Fibers.
J.Mol.Biol., 435, 2023
|
|
6RZU
 
 | | Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (14.7 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
8J22
 
 | | Cryo-EM structure of FFAR2 complex bound with TUG-1375 | | Descriptor: | (2R,4R)-2-(2-chlorophenyl)-3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)phenyl]carbonyl-1,3-thiazolidine-4-carboxylic acid, Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
8J20
 
 | | Cryo-EM structure of FFAR3 bound with valeric acid and AR420626 | | Descriptor: | (4R)-N-[2,5-bis(chloranyl)phenyl]-4-(furan-2-yl)-2-methyl-5-oxidanylidene-4,6,7,8-tetrahydro-1H-quinoline-3-carboxamide, Free fatty acid receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
8J24
 
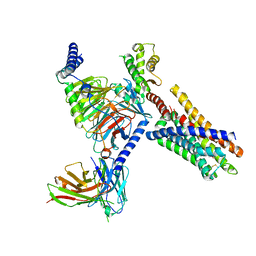 | | Cryo-EM structure of FFAR2 complex bound with acetic acid | | Descriptor: | ACETATE ION, Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tai, L, Li, F, Tang, W, Sun, X, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
8J21
 
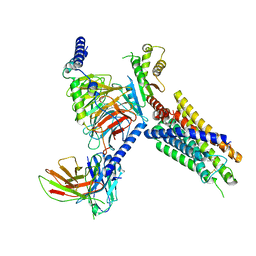 | | Cryo-EM structure of FFAR3 complex bound with butyrate acid | | Descriptor: | Free fatty acid receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
3W97
 
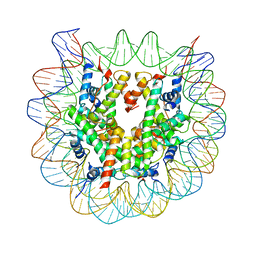 | | Crystal Structure of Human Nucleosome Core Particle lacking H2B N-terminal region | | Descriptor: | 146-mer DNA, Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Iwasaki, W, Miya, Y, Horikoshi, N, Osakabe, A, Tachiwana, H, Shibata, T, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2013-04-01 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Contribution of histone N-terminal tails to the structure and stability of nucleosomes
FEBS Open Bio, 3, 2013
|
|
8JJN
 
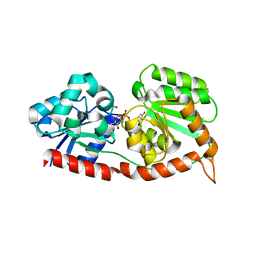 | |
4Y2I
 
 | | Gold ion bound to GolB | | Descriptor: | GOLD ION, Putative metal-binding transport protein | | Authors: | Wei, W, Wang, F, Ma, L, Zhao, J. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights and the Surprisingly Low Mechanical Stability of the Au-S Bond in the Gold-Specific Protein GolB
J.Am.Chem.Soc., 137, 2015
|
|
4Y07
 
 | | Crystal structure of the HECT domain of human WWP2 | | Descriptor: | NEDD4-like E3 ubiquitin-protein ligase WWP2 | | Authors: | Gong, W, Li, J, Li, Z, Xu, Y. | | Deposit date: | 2015-02-05 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | Structure of the HECT domain of human WWP2
Acta Crystallogr.,Sect.F, 71, 2015
|
|
8J1I
 
 | | Crystal Structure of EphA8/SASH1 Complex | | Descriptor: | Ephrin type-A receptor 8, SAM and SH3 domain-containing protein 1 | | Authors: | Liu, W, Li, J, Ding, Y. | | Deposit date: | 2023-04-12 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of EphA8 and SASH1 complex at 1.60 Angstroms resolution
To Be Published
|
|
4Y2K
 
 | | reduced form of apo-GolB | | Descriptor: | Putative metal-binding transport protein | | Authors: | Wei, W, Wang, F, Ma, L, Zhao, J. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights and the Surprisingly Low Mechanical Stability of the Au-S Bond in the Gold-Specific Protein GolB
J.Am.Chem.Soc., 137, 2015
|
|
8JIX
 
 | |
3OFU
 
 | | Crystal Structure of Cytochrome P450 CYP101C1 | | Descriptor: | (3E)-4-(2,6,6-trimethylcyclohex-1-en-1-yl)but-3-en-2-one, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhou, W, Ma, M, Bell, S.G, Yang, W, Hao, Y, Rees, N.H, Bartlam, M, Wong, L.-L, Rao, Z. | | Deposit date: | 2010-08-16 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Analysis of CYP101C1 from Novosphingobium aromaticivorans DSM12444.
Chembiochem, 12, 2011
|
|
8B7Z
 
 | | Bacterial chalcone isomerase H33A with taxifolin | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Palm, G.J, Hinrichs, W. | | Deposit date: | 2022-10-03 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for (2 R ,3 R )-Taxifolin Binding and Reaction Products to the Bacterial Chalcone Isomerase of Eubacterium ramulus .
Molecules, 27, 2022
|
|
8B7R
 
 | | Bacterial chalcone isomerase with taxifolin chalcone | | Descriptor: | (Z)-3-[3,4-bis(oxidanyl)phenyl]-2-oxidanyl-1-[2,4,6-tris(oxidanyl)phenyl]prop-2-en-1-one, CHLORIDE ION, Chalcone isomerase, ... | | Authors: | Hinrichs, W, Palm, G.J. | | Deposit date: | 2022-10-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for (2 R ,3 R )-Taxifolin Binding and Reaction Products to the Bacterial Chalcone Isomerase of Eubacterium ramulus .
Molecules, 27, 2022
|
|
8B7U
 
 | | Bacterial chalcone isomerase H33A with taxifolin | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, CHLORIDE ION, Chalcone isomerase, ... | | Authors: | Hinrichs, W, Palm, G.J. | | Deposit date: | 2022-10-03 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for (2 R ,3 R )-Taxifolin Binding and Reaction Products to the Bacterial Chalcone Isomerase of Eubacterium ramulus .
Molecules, 27, 2022
|
|
1U4E
 
 | | Crystal Structure of Cytoplasmic Domains of GIRK1 channel | | Descriptor: | G protein-activated inward rectifier potassium channel 1 | | Authors: | Pegan, S, Arrabit, C, Zhou, W, Kwiatkowski, W, Slesinger, P.A, Choe, S. | | Deposit date: | 2004-07-24 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Cytoplasmic domain structures of Kir2.1 and Kir3.1 show sites for modulating gating and rectification
Nat.Neurosci., 8, 2005
|
|
2M1D
 
 | |
3EQY
 
 | |
8JHH
 
 | | Glycoside hydrolase family 55 endo-beta-1,3-glucanase from Microdochium nivale | | Descriptor: | GLYCEROL, MnLam55A | | Authors: | Ota, T, Saburi, W, Yamashita, K, Tagami, T, Yu, J, Komba, S, Jewell, L.E, Hsiang, T, Imai, R, Yao, M, Mori, H. | | Deposit date: | 2023-05-23 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular mechanism for endo-type action of glycoside hydrolase family 55 endo-beta-1,3-glucanase on beta 1-3/1-6-glucan.
J.Biol.Chem., 299, 2023
|
|
