7GIQ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BEN-DND-c852c98b-5 (Mpro-P0124) | | Descriptor: | (4R)-6-chloro-N-[6-(methanesulfonyl)isoquinolin-4-yl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.916 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GNL
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with NIR-WEI-dcc3321b-1 (Mpro-P2757) | | Descriptor: | (4S)-6-chloro-2-{2-[4-(4-ethylpiperazin-1-yl)anilino]-2-oxoethyl}-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.681 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GHR
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-ce760d3f-8 (Mpro-P0017) | | Descriptor: | (3S)-3-(4-chlorophenyl)-1-(isoquinolin-4-yl)piperidin-2-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GJ5
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDG-MED-5d232de5-5 (Mpro-P0171) | | Descriptor: | (4R)-6-chloro-N-(isoquinolin-4-yl)-1-methyl-1,2,3,4-tetrahydroquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
3KO1
 
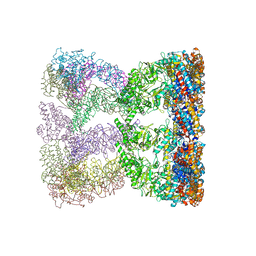 | | Cystal structure of thermosome from Acidianus tengchongensis strain S5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin | | Authors: | Huo, Y, Zhang, K, Hu, Z, Wang, L, Zhai, Y, Zhou, Q, Lander, G, He, Y, Zhu, J, Xu, W, Dong, Z, Sun, F. | | Deposit date: | 2009-11-12 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of group II chaperonin in the open state.
Structure, 18, 2010
|
|
3ZOI
 
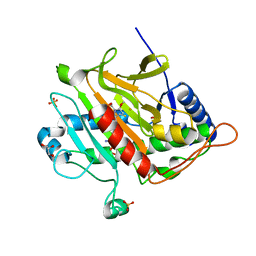 | | ISOPENICILLIN N SYNTHASE WITH AC-O-METHYL-D-THREONINE | | Descriptor: | DELTA-(L-ALPHA-AMINOADIPOYL)-L-CYSTEINYL-O-METHYL-D-THREONINE, FE (III) ION, GLYCEROL, ... | | Authors: | Rutledge, P.J, Clifton, I.J, Ge, W. | | Deposit date: | 2013-02-21 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Crystal Structure of an Isopenicillin N Synthase Complex with an Ethereal Substrate Analogue Reveals Water in the Oxygen Binding Site.
FEBS Lett., 587, 2013
|
|
7GJL
 
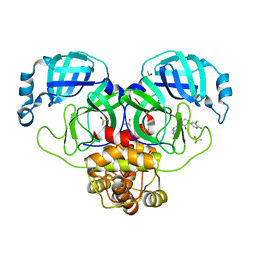 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-UNI-3735e77e-1 (Mpro-P0600) | | Descriptor: | (3R)-3-(3,4-dichlorophenyl)-1-(isoquinolin-4-yl)piperidin-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
2PXX
 
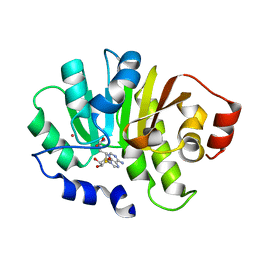 | | Human putative methyltransferase MGC2408 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, UNKNOWN ATOM OR ION, Uncharacterized protein MGC2408 | | Authors: | Tempel, W, Wu, H, Dombrovsky, L, Zeng, H, Zhu, H, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-14 | | Release date: | 2007-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An intact SAM-dependent methyltransferase fold is encoded by the human endothelin-converting enzyme-2 gene.
Proteins, 74, 2009
|
|
7GI6
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with RAL-THA-4aa06b95-1 (Mpro-P0056) | | Descriptor: | (4R)-6-chloro-N~4~-(isoquinolin-4-yl)-3,4-dihydroquinoline-1,4(2H)-dicarboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GIL
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with RAL-THA-2d450e86-6 (Mpro-P0108) | | Descriptor: | 2-(3-chloro-4-fluorophenyl)-N-(isoquinolin-4-yl)acetamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GK0
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BEN-BAS-c2bc0d80-7 (Mpro-P0765) | | Descriptor: | (1'M,4S)-6-chloro-1'-(isoquinolin-4-yl)-3'-methyl-2,3-dihydrospiro[[1]benzopyran-4,4'-imidazolidine]-2',5'-dione, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GKJ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDG-MED-ba1ac7b9-19 (Mpro-P0904) | | Descriptor: | (4S)-6-chloro-N-(isoquinolin-4-yl)-4-{2-[(4R,8S)-8-methyl-5,6-dihydro[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl]-2-oxoethyl}-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
2PYP
 
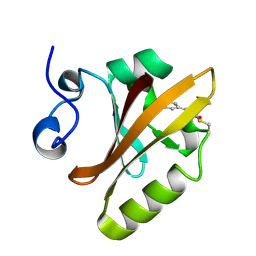 | | PHOTOACTIVE YELLOW PROTEIN, PHOTOSTATIONARY STATE, 50% GROUND STATE, 50% BLEACHED | | Descriptor: | Photoactive yellow protein | | Authors: | Genick, U.K, Borgstahl, G.E.O, Ng, K, Ren, Z, Pradervand, C, Burke, P, Srajer, V, Teng, T, Schildkamp, W, Mcree, D.E, Moffat, K, Getzoff, E.D. | | Deposit date: | 1997-02-03 | | Release date: | 1998-04-29 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a protein photocycle intermediate by millisecond time-resolved crystallography.
Science, 275, 1997
|
|
3QFO
 
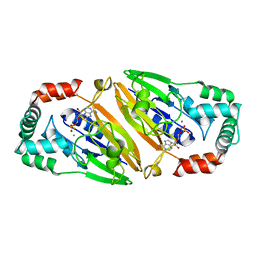 | | Crystal structure of Streptococcal asymmetric Ap4A hydrolase and phosphodiesterase Spr1479/SapH im complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, FE (III) ION, MANGANESE (II) ION, ... | | Authors: | Jiang, Y.L, Zhang, J.W, Yu, W.L, Cheng, W, Zhang, C.C, Zhou, C.Z, Chen, Y. | | Deposit date: | 2011-01-22 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and enzymatic characterization of a Streptococcal ATP/diadenosine polyphosphate and phosphodiester hydrolase Spr1479/SapH
To be Published
|
|
3Q9L
 
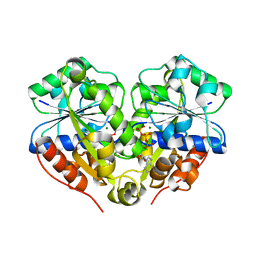 | | The structure of the dimeric E.coli MinD-ATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Septum site-determining protein minD | | Authors: | Wu, W, Park, K.-T, Lutkenhaus, J, Holyoak, T. | | Deposit date: | 2011-01-08 | | Release date: | 2011-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Determination of the structure of the MinD-ATP complex reveals the orientation of MinD on the membrane and the relative location of the binding sites for MinE and MinC.
Mol.Microbiol., 79, 2011
|
|
6EMT
 
 | |
6EMV
 
 | |
3QBO
 
 | | Crystal structure of phosphoserine aminotransferase from Yersinia pestis CO92 | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Phosphoserine aminotransferase | | Authors: | Nocek, B, Maltseva, N, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-13 | | Release date: | 2011-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structure of phosphoserine aminotransferase from Yersinia pestis CO92
TO BE PUBLISHED
|
|
3KUQ
 
 | | Crystal structure of the DLC1 RhoGAP domain | | Descriptor: | Rho GTPase-activating protein 7, UNKNOWN ATOM OR ION | | Authors: | Nedyalkova, L, Tempel, W, Tong, Y, MacKenzie, F, Shen, L, Zhong, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-27 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the DLC1 RhoGAP domain
to be published
|
|
5WYG
 
 | | The crystal structure of the apo form of Mtb MazF | | Descriptor: | Probable endoribonuclease MazF7 | | Authors: | Xie, W, Chen, R, Tu, J. | | Deposit date: | 2017-01-13 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.356 Å) | | Cite: | Structure of the MazF-mt9 toxin, a tRNA-specific endonuclease from Mycobacterium tuberculosis
Biochem. Biophys. Res. Commun., 486, 2017
|
|
5WZ2
 
 | | Crystal structure of Zika virus NS5 methyltransferase bound to SAM and RNA analogue (m7GpppA) | | Descriptor: | NS5 MTase, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, S-ADENOSYLMETHIONINE | | Authors: | Duan, W, Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of Zika virus NS5 reveals conserved drug targets.
EMBO J., 36, 2017
|
|
3NI7
 
 | | Crystal structure of the TetR transcriptional regulator from Nitrosomonas europaea ATCC 19718 | | Descriptor: | Bacterial regulatory proteins, TetR family | | Authors: | Knapik, A, Chruszcz, M, Cymborowski, M, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-15 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of the TetR transcriptional regulator from Nitrosomonas europaea ATCC 19718
To be Published
|
|
5WVN
 
 | | Crystal structure of MBS-BaeS fusion protein | | Descriptor: | Maltose-binding periplasmic protein,Two-component system sensor kinase, SULFATE ION | | Authors: | Wang, W, Zhang, Y, Ran, T, Xu, D. | | Deposit date: | 2016-12-26 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the sensor domain of BaeS from Serratia marcescens FS14
Proteins, 85, 2017
|
|
2RLO
 
 | | Split PH domain of PI3-kinase enhancer | | Descriptor: | Centaurin-gamma 1 | | Authors: | Wen, W, Zhang, M. | | Deposit date: | 2007-07-21 | | Release date: | 2008-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Split pleckstrin homology domain-mediated cytoplasmic-nuclear localization of PI3-kinase enhancer GTPase
J.Mol.Biol., 378, 2008
|
|
3L19
 
 | | Crystal structure of calcium binding domain of CpCDPK3, cgd5_820 | | Descriptor: | CALCIUM ION, Calcium/calmodulin dependent protein kinase with a kinase domain and 4 calmodulin like EF hands, GLYCEROL, ... | | Authors: | Qiu, W, Hutchinson, A, Wernimont, A, Walker, J.R, Sullivan, H, Lin, Y.-H, Mackenzie, F, Kozieradzki, I, Cossar, D, Schapira, M, Senisterra, G, Vedadi, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Bochkarev, A, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-12-11 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of calcium binding domain of CpCDPK3, cgd5_820
To be Published
|
|
