1L1Q
 
 | | Crystal Structure of APRTase from Giardia lamblia Complexed with 9-deazaadenine | | Descriptor: | 9-DEAZAADENINE, Adenine phosphoribosyltransferase, SULFATE ION | | Authors: | Shi, W, Sarver, A.E, Wang, C.C, Tanaka, K.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2002-02-19 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Closed Site Complexes of Adenine Phosphoribosyltransferase from Giardia lamblia
Reveal a Mechanism of Ribosyl Migration.
J.Biol.Chem., 277, 2002
|
|
6BKZ
 
 | | Novel Modes of Inhibition of Wild-Type IDH1: Non-equivalent Allosteric Inhibition with Cmpd3 | | Descriptor: | (7R)-1-[(4-fluorophenyl)methyl]-N-{3-[(1R)-1-hydroxyethyl]phenyl}-7-methyl-5-(1H-pyrrole-2-carbonyl)-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridine-3-carboxamide, Isocitrate dehydrogenase [NADP] cytoplasmic, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jakob, C.G, Qiu, W. | | Deposit date: | 2017-11-09 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Novel Modes of Inhibition of Wild-Type Isocitrate Dehydrogenase 1 (IDH1): Direct Covalent Modification of His315.
J. Med. Chem., 61, 2018
|
|
6BM4
 
 | | Pol II elongation complex with an abasic lesion at i-1 position,soaking UMPNPP | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, DNA (5'-D(P*CP*AP*(3DR)P*CP*TP*CP*TP*TP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, W, Wang, D. | | Deposit date: | 2017-11-13 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Structural basis of transcriptional stalling and bypass of abasic DNA lesion by RNA polymerase II.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1BMV
 
 | | PROTEIN-RNA INTERACTIONS IN AN ICOSAHEDRAL VIRUS AT 3.0 ANGSTROMS RESOLUTION | | Descriptor: | PROTEIN (ICOSAHEDRAL VIRUS - A DOMAIN), PROTEIN (ICOSAHEDRAL VIRUS - B AND C DOMAIN), RNA (5'-R(*GP*GP*UP*CP*AP*AP*AP*AP*UP*GP*C)-3') | | Authors: | Chen, Z, Stauffacher, C, Li, Y, Schmidt, T, Bomu, W, Kamer, G, Shanks, M, Lomonossoff, G, Johnson, J.E. | | Deposit date: | 1989-10-09 | | Release date: | 1989-10-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Protein-RNA interactions in an icosahedral virus at 3.0 A resolution.
Science, 245, 1989
|
|
2QYP
 
 | |
1L2N
 
 | | Smt3 Solution Structure | | Descriptor: | Ubiquitin-like protein SMT3 | | Authors: | Sheng, W, Liao, X. | | Deposit date: | 2002-02-22 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a yeast ubiquitin-like protein Smt3: the role of structurally less defined sequences in protein-protein recognitions.
Protein Sci., 11, 2002
|
|
3JWP
 
 | | Crystal structure of Plasmodium falciparum SIR2A (PF13_0152) in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, TRIETHYLENE GLYCOL, Transcriptional regulatory protein sir2 homologue, ... | | Authors: | Wernimont, A.K, Hutchinson, A, Lin, Y.H, MacKenzie, F, Senisterra, G, Allali-Hassanali, A, Vedadi, M, Ravichandran, M, Cossar, D, Kozieradzki, I, Zhao, Y, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Qiu, W, Brand, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-18 | | Release date: | 2009-10-20 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of Plasmodium falciparum SIR2A (PF13_0152) in complex with AMP
TO BE PUBLISHED
|
|
3ODU
 
 | | The 2.5 A structure of the CXCR4 chemokine receptor in complex with small molecule antagonist IT1t | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, ... | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-11 | | Release date: | 2010-10-27 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3K9P
 
 | | The crystal structure of E2-25K and ubiquitin complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 K | | Authors: | Kang, G.B, Ko, S, Song, S.M, Lee, W, Eom, S.H. | | Deposit date: | 2009-10-16 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of E2-25K/UBB+1 interaction leading to proteasome inhibition and neurotoxicity
J.Biol.Chem., 285, 2010
|
|
5H6U
 
 | | Structure of alginate-binding protein AlgQ2 in complex with an alginate pentasaccharide | | Descriptor: | AlgQ2, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Uenishi, K, Kaneko, A, Maruyama, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2016-11-15 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | A solute-binding protein in the closed conformation induces ATP hydrolysis in a bacterial ATP-binding cassette transporter involved in the import of alginate
J. Biol. Chem., 292, 2017
|
|
1L2Q
 
 | | Crystal Structure of the Methanosarcina barkeri Monomethylamine Methyltransferase (MtmB) | | Descriptor: | AMMONIUM ION, monomethylamine methyltransferase | | Authors: | Hao, B, Gong, W, Ferguson, T.K, James, C.M, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2002-02-24 | | Release date: | 2002-06-05 | | Last modified: | 2014-08-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A new UAG-encoded residue in the structure of a methanogen methyltransferase.
Science, 296, 2002
|
|
6BSG
 
 | | Structure of HIV-1 RT complexed with RNA/DNA hybrid in an RNA hydrolysis-off mode | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tian, L, Kim, M, Yang, W. | | Deposit date: | 2017-12-03 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5HJK
 
 | |
1BQB
 
 | | AUREOLYSIN, STAPHYLOCOCCUS AUREUS METALLOPROTEINASE | | Descriptor: | CALCIUM ION, PROTEIN (AUREOLYSIN), ZINC ION | | Authors: | Medrano, F.J, Banbula, A, Potempa, J, Travis, J, Bode, W. | | Deposit date: | 1998-07-14 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Amino-acid sequence and three-dimensional structure of the Staphylococcus aureus metalloproteinase at 1.72 A resolution.
Structure, 6, 1998
|
|
7EVM
 
 | | Cryo-EM structure of the compound 2-bound human GLP-1 receptor-Gs complex | | Descriptor: | CHOLESTEROL, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z, Chen, L, Ma, H, Zhou, Q, Zou, X, Ye, C, Dai, A, Liu, Q, Huang, W, Sun, X, Wang, X, Xu, P, Zhao, L, Xia, T, Zhong, W, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2021-05-21 | | Release date: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor.
Nat Commun, 12, 2021
|
|
3K5M
 
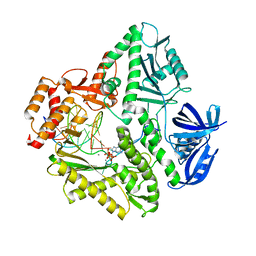 | | Crystal structure of E.coli Pol II-abasic DNA-ddGTP Lt(-2, 2) ternary complex | | Descriptor: | 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*AP*GP*TP*CP*CP*TP*GP*(3DR)P*AP*CP*GP*CP*TP*AP*GP*GP*CP*AP*CP*A)-3'), ... | | Authors: | Yang, W, Wang, F. | | Deposit date: | 2009-10-07 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural insight into translesion synthesis by DNA Pol II.
Cell(Cambridge,Mass.), 139, 2009
|
|
1L1R
 
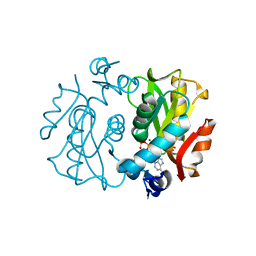 | | Crystal Structure of APRTase from Giardia lamblia Complexed with 9-deazaadenine, Mg2+ and PRPP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 9-DEAZAADENINE, Adenine phosphoribosyltransferase, ... | | Authors: | Shi, W, Sarver, A.E, Wang, C.C, Tanaka, K.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2002-02-19 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Closed Site Complexes of Adenine Phosphoribosyltransferase from Giardia lamblia
Reveal a Mechanism of Ribosyl Migration.
J.Biol.Chem., 277, 2002
|
|
3OPK
 
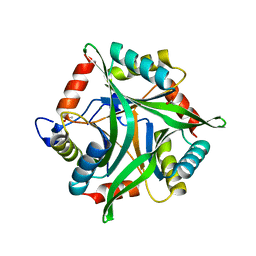 | | Crystal structure of divalent-cation tolerance protein CutA from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | | Descriptor: | ACETATE ION, Divalent-cation tolerance protein cutA, MAGNESIUM ION, ... | | Authors: | Nocek, B, Mulligan, R, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-01 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of divalent-cation tolerance protein CutA from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2
TO BE PUBLISHED
|
|
1ITF
 
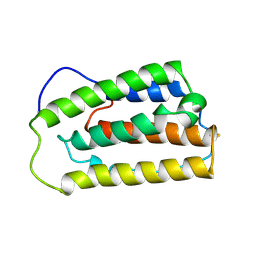 | | INTERFERON ALPHA-2A, NMR, 24 STRUCTURES | | Descriptor: | INTERFERON ALPHA-2A | | Authors: | Klaus, W, Gsell, B, Labhardt, A.M, Wipf, B, Senn, H. | | Deposit date: | 1997-08-22 | | Release date: | 1997-12-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional high resolution structure of human interferon alpha-2a determined by heteronuclear NMR spectroscopy in solution.
J.Mol.Biol., 274, 1997
|
|
3OQO
 
 | | Ccpa-hpr-ser46p-syn cre | | Descriptor: | 5'-D(*CP*TP*GP*AP*AP*AP*GP*CP*GP*CP*TP*AP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*TP*AP*GP*CP*GP*CP*TP*TP*TP*CP*AP*G)-3', Catabolite control protein A, ... | | Authors: | schumacher, M.A, Sprehe, M, Bartholomae, M, Hillen, W, Brennan, R.G. | | Deposit date: | 2010-09-03 | | Release date: | 2011-10-26 | | Last modified: | 2014-04-09 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structures of carbon catabolite protein A-(HPr-Ser46-P) bound to diverse catabolite response element sites reveal the basis for high-affinity binding to degenerate DNA operators.
Nucleic Acids Res., 39, 2011
|
|
1L7L
 
 | | Crystal structure of Pseudomonas aeruginosa lectin 1 determined by single wavelength anomalous scattering phasing method | | Descriptor: | CALCIUM ION, PA-I galactophilic lectin | | Authors: | Liu, Z.J, Tempel, W, Lin, D, Karaveg, K, Doyle, R.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2002-03-15 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure determination of P. aeruginosa lectin-1 using single
wavelength anomalous scattering data from native crystals (P028)
AM.CRYST.ASSOC.,ABSTR.PAPERS (ANNUAL MEETING), 29, 2002
|
|
3SWH
 
 | | Munc13-1, MUN domain, C-terminal module | | Descriptor: | Protein unc-13 homolog A | | Authors: | Tomchick, D.R, Rizo, J, Li, W. | | Deposit date: | 2011-07-13 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Crystal Structure of a Munc13 C-terminal Module Exhibits a Remarkable Similarity to Vesicle Tethering Factors.
Structure, 19, 2011
|
|
3J45
 
 | | Structure of a non-translocating SecY protein channel with the 70S ribosome | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L23, 50S ribosomal protein L24, ... | | Authors: | Menetret, J.F, Park, E, Gumbart, J.C, Ludtke, S.J, Li, W, Whynot, A, Rapoport, T.A, Akey, C.W. | | Deposit date: | 2013-06-18 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structure of the SecY channel during initiation of protein translocation.
Nature, 506, 2013
|
|
6AOZ
 
 | |
3OWC
 
 | | Crystal structure of GNAT superfamily protein PA2578 from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, Probable acetyltransferase | | Authors: | Majorek, K.A, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-09-17 | | Release date: | 2010-11-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of GNAT superfamily protein PA2578 from Pseudomonas aeruginosa
To be Published
|
|
