7M83
 
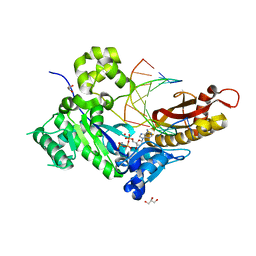 | | Human DNA Pol eta S113A with dA-ended primer and dATP: in crystallo reaction for 0 s | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*A)-3'), ... | | Authors: | Gregory, M.T, Gao, Y, Yang, W. | | Deposit date: | 2021-03-29 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Multiple deprotonation paths of the nucleophile 3'-OH in the DNA synthesis reaction.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7M7L
 
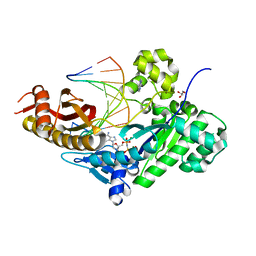 | | Human DNA Pol eta with dA-ended primer and dAMPNPP | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*A)-3'), DNA (5'-D(*CP*AP*TP*TP*TP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Gregory, M.T, Gao, Y, Yang, W. | | Deposit date: | 2021-03-28 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Multiple deprotonation paths of the nucleophile 3'-OH in the DNA synthesis reaction.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7M7O
 
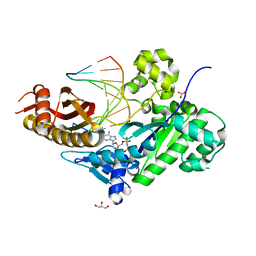 | | Human DNA Pol eta with dT-ended primer and 0.1 mM dAMPNPP | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Gregory, M.T, Gao, Y, Yang, W. | | Deposit date: | 2021-03-28 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Multiple deprotonation paths of the nucleophile 3'-OH in the DNA synthesis reaction.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7M7U
 
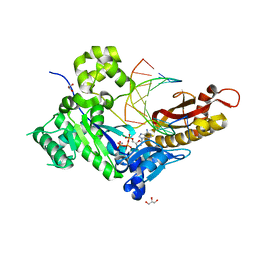 | | Human DNA Pol eta S113A with dT-ended primer and dATP: in crystallo reaction for 480s | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Gregory, M.T, Gao, Y, Yang, W. | | Deposit date: | 2021-03-28 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Multiple deprotonation paths of the nucleophile 3'-OH in the DNA synthesis reaction.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7M80
 
 | | Human DNA Pol eta with dA-ended primer and dATP: in crystallo reaction for 100 s | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Gregory, M.T, Gao, Y, Yang, W. | | Deposit date: | 2021-03-29 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Multiple deprotonation paths of the nucleophile 3'-OH in the DNA synthesis reaction.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7M8A
 
 | | Human DNA Pol eta S113A with rA-ended primer and dATP: in crystallo reaction for 40 s | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Gregory, M.T, Gao, Y, Yang, W. | | Deposit date: | 2021-03-29 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Multiple deprotonation paths of the nucleophile 3'-OH in the DNA synthesis reaction.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7M7P
 
 | | Human DNA Pol eta S113A with dA-ended primer and dAMPNPP | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*CP*AP*TP*TP*TP*TP*GP*AP*CP*GP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*CP*GP*TP*CP*AP*A)-3'), ... | | Authors: | Gregory, M.T, Gao, Y, Yang, W. | | Deposit date: | 2021-03-28 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Multiple deprotonation paths of the nucleophile 3'-OH in the DNA synthesis reaction.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7M84
 
 | | Human DNA Pol eta S113A with dA-ended primer and dATP: in crystallo reaction for 40 s | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*A)-3'), ... | | Authors: | Gregory, M.T, Gao, Y, Yang, W. | | Deposit date: | 2021-03-29 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Multiple deprotonation paths of the nucleophile 3'-OH in the DNA synthesis reaction.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7M86
 
 | | Human DNA Pol eta S113A with dA-ended primer and dATP: in crystallo reaction for 140 s | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Gregory, M.T, Gao, Y, Yang, W. | | Deposit date: | 2021-03-29 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Multiple deprotonation paths of the nucleophile 3'-OH in the DNA synthesis reaction.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7M8C
 
 | | Human DNA Pol eta S113A with rA-ended primer and dATP: in crystallo reaction for 230 s | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Gregory, M.T, Gao, Y, Yang, W. | | Deposit date: | 2021-03-29 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Multiple deprotonation paths of the nucleophile 3'-OH in the DNA synthesis reaction.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MBR
 
 | | Cryo-EM structure of zebrafish TRPM5 in the presence of 6 uM calcium (apo state) | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MBS
 
 | | Cryo-EM structure of zebrafish TRPM5 in the presence of 6 uM calcium (open state) | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MBT
 
 | | Cryo-EM structure of zebrafish TRPM5 E337A mutant in the presence of 5 mM calcium (low calcium occupancy in the transmembrane domain) | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MBQ
 
 | | Cryo-EM structure of zebrafish TRPM5 in the presence of 5 mM calcium | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MOY
 
 | | Structure of HDAC2 in complex with an inhibitor (compound 19) | | Descriptor: | (1S)-6-ethyl-N-{(1S)-1-[5-(2-ethyl-1-oxo-1,2-dihydroisoquinolin-6-yl)-1H-imidazol-2-yl]-7,7-dihydroxynonyl}-6-azaspiro[2.5]octane-1-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Discovery of macrocyclic HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
7MBU
 
 | | Cryo-EM structure of zebrafish TRPM5 E337A mutant in the presence of 5 mM calcium (high calcium occupancy in the transmembrane domain) | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MBV
 
 | | Cryo-EM structure of zebrafish TRPM5 in the presence of 5 mM calcium and 0.5 mM NDNA | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MOT
 
 | | Structure of HDAC2 in complex with an inhibitor (compound 9) | | Descriptor: | 5-{(1S)-7,7-dihydroxy-1-[(1-methylazetidine-3-carbonyl)amino]nonyl}-2-phenyl-1H-imidazole-4-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of macrocyclic HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
7MOS
 
 | | Structure of HDAC2 in complex with a macrocyclic inhibitor (compound 4) | | Descriptor: | (3S,18S,20aR)-18-(6,6-dihydroxyoctyl)-1,5,6,7,8,18,19,20a-octahydro-4H-14,17-epiminoazeto[1,2-g][1,7,10,13]benzoxatriazacycloheptadecin-20(2H)-one, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Discovery of macrocyclic HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
7MOX
 
 | | Structure of HDAC2 in complex with an inhibitor (compound 14) | | Descriptor: | (1S)-N-[(1S)-7,7-dihydroxy-1-{4-[(1R,4S)-1,2,3,4-tetrahydro-1,4-methanonaphthalen-6-yl]-1H-imidazol-2-yl}nonyl]-6-methyl-6-azaspiro[2.5]octane-1-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of macrocyclic HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
7MOZ
 
 | | Structure of HDAC2 in complex with a macrocyclic inhibitor (compound 25) | | Descriptor: | (1R,3S,6S,18R,27R)-6-(6,6-dihydroxyoctyl)-5,8,18,27,34-pentaazahexacyclo[25.2.2.1~7,10~.1~11,15~.1~14,18~.0~1,3~]tetratriaconta-7,9,11(33),12,14,16-hexaene-4,32-dione (non-preferred name), CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | Discovery of macrocyclic HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
6UIX
 
 | | Cryo-EM structure of human CALHM2 gap junction | | Descriptor: | Calcium homeostasis modulator protein 2 | | Authors: | Lu, W, Du, J, Choi, W. | | Deposit date: | 2019-10-01 | | Release date: | 2019-11-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structures and gating mechanism of human calcium homeostasis modulator 2.
Nature, 576, 2019
|
|
7MSY
 
 | | Structure of CalU17 from the Calicheamicin Biosynthesis Pathway of Micromonospora echinospora | | Descriptor: | CALCIUM ION, CHLORIDE ION, CalU17, ... | | Authors: | Kosgei, A.J, Miller, M.D, Xu, W, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2021-05-12 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The crystal structure of DynF from the dynemicin-biosynthesis pathway of Micromonospora chersina.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7ML6
 
 | | Structure of CalU17 from the Calicheamicin Biosynthesis Pathway of Micromonospora echinospora | | Descriptor: | CalU17, GLYCEROL | | Authors: | Kosgei, A.J, Miller, M.D, Xu, W, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2021-04-27 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of DynF from the dynemicin-biosynthesis pathway of Micromonospora chersina.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7MDW
 
 | |
