7TXT
 
 | | Structure of human serotonin transporter bound to small molecule '8090 in lipid nanodisc and NaCl | | Descriptor: | 1-[4-(4-fluorophenyl)-1,3-thiazol-2-yl]piperazine, 15B8 Fab heavy chain, 15B8 Fab light chain, ... | | Authors: | Singh, I, Seth, A, Billesboelle, C.B, Braz, J, Rodriguiz, R.M, Roy, K, Bekele, B, Craik, V, Huang, X.P, Boytsov, D, Lak, P, O'Donnell, H, Sandtner, W, Roth, B.L, Basbaum, A.I, Wetsel, W.C, Manglik, A, Shoichet, B.K, Rudnick, G. | | Deposit date: | 2022-02-09 | | Release date: | 2023-03-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-based discovery of conformationally selective inhibitors of the serotonin transporter.
Cell, 186, 2023
|
|
6A6R
 
 | | Crystal structure of the modified fructosyl peptide oxidase from Aspergillus nidulans, Seleno-methionine Derivative | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine: oxygen oxidoreductase, ... | | Authors: | Ogawa, N, Maruyama, Y, Itoh, T, Hashimoto, W, Murata, K. | | Deposit date: | 2018-06-29 | | Release date: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Creation of haemoglobin A1c direct oxidase from fructosyl peptide oxidase by combined structure-based site specific mutagenesis and random mutagenesis.
Sci Rep, 9, 2019
|
|
3LJU
 
 | | Crystal structure of full length centaurin alpha-1 bound with the head group of PIP3 | | Descriptor: | (2R)-3-{[(R)-{[(1S,2S,3R,4S,5S,6S)-2,6-dihydroxy-3,4,5-tris(phosphonooxy)cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}propane -1,2-diyl dioctanoate, Arf-GAP with dual PH domain-containing protein 1, ZINC ION | | Authors: | Shen, L, Tong, Y, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-26 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Phosphorylation-independent dual-site binding of the FHA domain of KIF13 mediates phosphoinositide transport via centaurin alpha1.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6ACD
 
 | |
3T2G
 
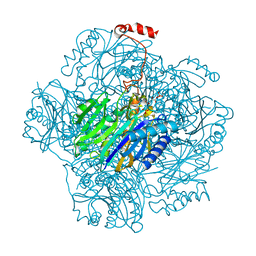 | | Fructose-1,6-bisphosphate aldolase/phosphatase from Thermoproteus neutrophilus, Y229F variant with DHAP | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-1,6-bisphosphate aldolase/phosphatase, MAGNESIUM ION | | Authors: | Du, J, Say, R, Lue, W, Fuchs, G, Einsle, O. | | Deposit date: | 2011-07-22 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Active-site remodelling in the bifunctional fructose-1,6-bisphosphate aldolase/phosphatase.
Nature, 478, 2011
|
|
1C6R
 
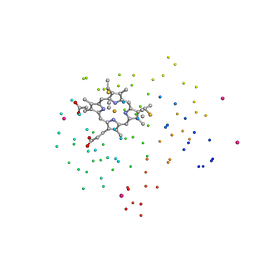 | | CRYSTAL STRUCTURE OF REDUCED CYTOCHROME C6 FROM THE GREEN ALGAE SCENEDESMUS OBLIQUUS | | Descriptor: | CADMIUM ION, CYTOCHROME C6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Schnackenberg, J, Than, M.E, Mann, K, Wiegand, G, Huber, R, Reuter, W. | | Deposit date: | 1999-04-06 | | Release date: | 2000-04-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Amino acid sequence, crystallization and structure determination of reduced and oxidized cytochrome c6 from the green alga Scenedesmus obliquus.
J.Mol.Biol., 290, 1999
|
|
2R2Q
 
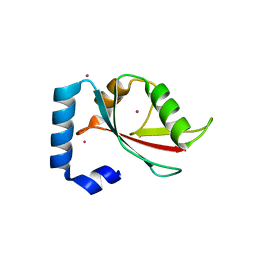 | | Crystal structure of human Gamma-Aminobutyric Acid Receptor-Associated Protein-like 1 (GABARAP1), Isoform CRA_a | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Gamma-aminobutyric acid receptor-associated protein-like 1, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Paramanathan, R, Davis, T, Mujib, S, Butler-Cole, C, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-08-27 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of human Gamma-Aminobutyric Acid Receptor-Associated Protein-like 1 (GABARAP1), Isoform CRA_a.
To be Published
|
|
1MRO
 
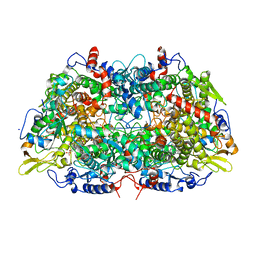 | | METHYL-COENZYME M REDUCTASE | | Descriptor: | 1-THIOETHANESULFONIC ACID, Coenzyme B, FACTOR 430, ... | | Authors: | Ermler, U, Grabarse, W. | | Deposit date: | 1997-10-01 | | Release date: | 1998-11-11 | | Last modified: | 2017-03-29 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystal structure of methyl-coenzyme M reductase: the key enzyme of biological methane formation.
Science, 278, 1997
|
|
3LPC
 
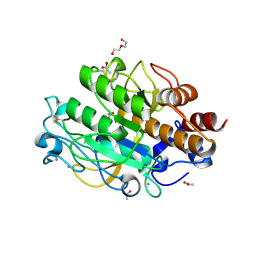 | | Crystal structure of a subtilisin-like protease | | Descriptor: | ACETATE ION, AprB2, CALCIUM ION, ... | | Authors: | Porter, C.J, Wong, W, Whisstock, J.C, Rood, J.I, Kennan, R.M. | | Deposit date: | 2010-02-05 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Subtilisin-Like Protease AprV2 Is Required for Virulence and Uses a Novel Disulphide-Tethered Exosite to Bind Substrates
Plos Pathog., 6, 2010
|
|
1H7S
 
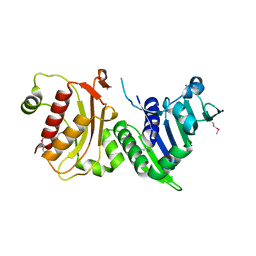 | | N-terminal 40kDa fragment of human PMS2 | | Descriptor: | PMS1 PROTEIN HOMOLOG 2 | | Authors: | Guarne, A, Junop, M.S, Yang, W. | | Deposit date: | 2001-07-10 | | Release date: | 2001-11-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Function of the N-Terminal 40 kDa Fragment of Human Pms2: A Monomeric Ghl ATPase
Embo J., 20, 2001
|
|
6ACC
 
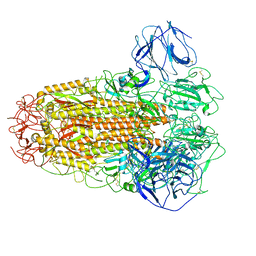 | |
1GTL
 
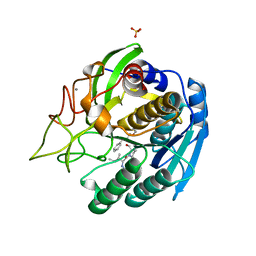 | | The thermostable serine-carboxyl type proteinase, kumamolisin (KSCP) - complex with Ac-Ile-Pro-Phe-cho | | Descriptor: | ALDEHYDE INHIBITOR, CALCIUM ION, KUMAMOLYSIN, ... | | Authors: | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | Deposit date: | 2002-01-16 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
1MWO
 
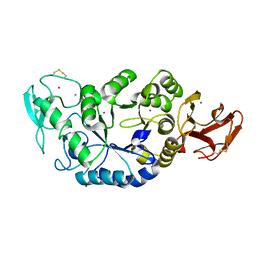 | | Crystal Structure Analysis of the Hyperthermostable Pyrocoocus woesei alpha-amylase | | Descriptor: | CALCIUM ION, ZINC ION, alpha amylase | | Authors: | Linden, A, Mayans, O, Meyer-Klaucke, W, Antranikian, G, Wilmanns, M. | | Deposit date: | 2002-09-30 | | Release date: | 2003-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Differential Regulation of a Hyperthermophilic alpha-Amylase with a Novel (Ca,Zn) Two-metal Center by Zinc
J.Biol.Chem., 278, 2003
|
|
3OQM
 
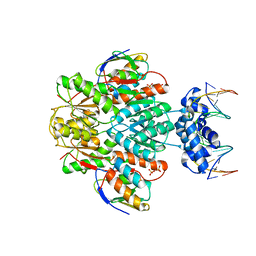 | | structure of ccpa-hpr-ser46p-ackA2 complex | | Descriptor: | 5'-D(*TP*TP*GP*AP*TP*AP*AP*CP*GP*CP*TP*TP*AP*CP*AP*A)-3', 5'-D(*TP*TP*GP*TP*AP*AP*GP*CP*GP*TP*TP*AP*TP*CP*AP*A)-3', Catabolite control protein A, ... | | Authors: | Schumacher, M.A, Sprehe, M, Bartholomae, M, Hillen, W, Brennan, R.G. | | Deposit date: | 2010-09-03 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structures of carbon catabolite protein A-(HPr-Ser46-P) bound to diverse catabolite response element sites reveal the basis for high-affinity binding to degenerate DNA operators.
Nucleic Acids Res., 39, 2011
|
|
3ORQ
 
 | | Crystal Structure of N5-Carboxyaminoimidazole synthetase from Staphylococcus aureus complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Brugarolas, P, Duguid, E.M, Zhang, W, Poor, C.B, He, C. | | Deposit date: | 2010-09-07 | | Release date: | 2011-07-20 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and biochemical characterization of N5-carboxyaminoimidazole ribonucleotide synthetase and N5-carboxyaminoimidazole ribonucleotide mutase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3OT1
 
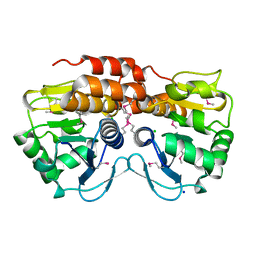 | | Crystal structure of VC2308 protein | | Descriptor: | 4-methyl-5(B-hydroxyethyl)-thiazole monophosphate biosynthesis enzyme, CHLORIDE ION, SODIUM ION | | Authors: | Niedzialkowska, E, Wawrzak, Z, Chruszcz, M, Porebski, P, Skarina, T, Huang, X, Grimshaw, S, Cymborowski, M, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-10 | | Release date: | 2010-09-22 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystal structure of VC2308 protein
To be Published
|
|
1N25
 
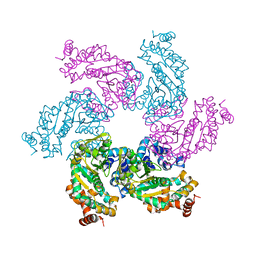 | | Crystal structure of the SV40 Large T antigen helicase domain | | Descriptor: | Large T Antigen, ZINC ION | | Authors: | Li, D, Zhao, R, Lilyestrom, W, Gai, D, Zhang, R, DeCaprio, J.A, Fanning, E, Jochimiak, A, Szakonyi, G, Chen, X.S. | | Deposit date: | 2002-10-21 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen
Nature, 423, 2003
|
|
2P0E
 
 | | Human nicotinamide riboside kinase 1 in complex with tiazofurin | | Descriptor: | (1R)-1-[4-(AMINOCARBONYL)-1,3-THIAZOL-2-YL]-1,4-ANHYDRO-D-RIBITOL, CHLORIDE ION, Nicotinamide riboside kinase 1, ... | | Authors: | Rabeh, W.M, Tempel, W, Nedyalkova, L, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Brenner, C, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-28 | | Release date: | 2007-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nicotinamide Riboside Kinase Structures Reveal New Pathways to NAD(+).
Plos Biol., 5, 2007
|
|
2P0M
 
 | | Revised structure of rabbit reticulocyte 15S-lipoxygenase | | Descriptor: | (2E)-3-(2-OCT-1-YN-1-YLPHENYL)ACRYLIC ACID, Arachidonate 15-lipoxygenase, FE (II) ION | | Authors: | Choi, J, Chon, J.K, Kim, S, Shin, W. | | Deposit date: | 2007-02-28 | | Release date: | 2007-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational flexibility in mammalian 15S-lipoxygenase: Reinterpretation of the crystallographic data.
Proteins, 70, 2008
|
|
1GT9
 
 | | High resolution crystal structure of a thermostable serine-carboxyl type proteinase, kumamolisin (kscp) | | Descriptor: | CALCIUM ION, KUMAMOLYSIN, SULFATE ION | | Authors: | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | Deposit date: | 2002-01-14 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
2P1U
 
 | | Crystal structure of the ligand binding domain of the retinoid X receptor alpha in complex with 3-(2'-ethoxy)-tetrahydronaphtyl cinnamic acid and a fragment of the coactivator TIF-2 | | Descriptor: | (2E)-3-[3-(3-ETHOXY-5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)-4-HYDROXYPHENYL]ACRYLIC ACID, Nuclear receptor coactivator 2 peptide, Retinoic acid receptor RXR-alpha | | Authors: | Bourguet, W, Nahoum, V. | | Deposit date: | 2007-03-06 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Modulators of the structural dynamics of the retinoid X receptor to reveal receptor function.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1MR1
 
 | | Crystal Structure of a Smad4-Ski Complex | | Descriptor: | Mothers against decapentaplegic homolog 4, Ski oncogene, ZINC ION | | Authors: | Wu, J.-W, Krawitz, A.R, Chai, J, Li, W, Zhang, F, Luo, K, Shi, Y. | | Deposit date: | 2002-09-17 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Mechanism of Smad4 Recognition by the Nuclear Oncoprotein Ski: Insights on Ski-mediated Repression of TGF-beta Signaling
Cell(Cambridge,Mass.), 111, 2002
|
|
2P3N
 
 | | Thermotoga maritima IMPase TM1415 | | Descriptor: | Inositol-1-monophosphatase, MAGNESIUM ION | | Authors: | Stieglitz, K.A, Roberts, M.F, Li, W, Stec, B. | | Deposit date: | 2007-03-09 | | Release date: | 2007-04-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the tetrameric inositol 1-phosphate phosphatase (TM1415) from the hyperthermophile, Thermotoga maritima.
Febs J., 274, 2007
|
|
7UXU
 
 | | CryoEM structure of the TIR domain from AbTir in complex with 3AD | | Descriptor: | Molecular chaperone Tir, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(8-azanylisoquinolin-2-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Li, S, Nanson, J.D, Manik, M.K, Gu, W, Landsberg, M.J, Ve, T, Kobe, B. | | Deposit date: | 2022-05-06 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
1N4O
 
 | |
