4U7U
 
 | | Crystal structure of RNA-guided immune Cascade complex from E.coli | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Zhao, H, Sheng, G, Wang, J, Wang, M, Bunkoczi, G, Gong, W, Wei, Z, Wang, Y. | | Deposit date: | 2014-07-31 | | Release date: | 2014-08-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli
Nature, 515, 2014
|
|
2MSX
 
 | | The solution structure of the MANEC-type domain from Hepatocyte Growth Factor Inhibitor 1 reveals an unexpected PAN/apple domain-type fold | | Descriptor: | Kunitz-type protease inhibitor 1 | | Authors: | Hong, Z, Nowakowski, M.E, Spronk, C, Petersen, S.V, Petersen, J.S, Kozminski, W, Mulder, F, Jensen, J.K. | | Deposit date: | 2014-08-09 | | Release date: | 2014-12-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the MANEC-type domain from hepatocyte growth factor activator inhibitor-1 reveals an unexpected PAN/apple domain-type fold.
Biochem.J., 466, 2015
|
|
1Z8Y
 
 | | Mapping the E2 Glycoprotein of Alphaviruses | | Descriptor: | Capsid protein C, Spike glycoprotein E1, Spike glycoprotein E2 | | Authors: | Mukhopadhyay, S, Zhang, W, Gabler, S, Chipman, P.R, Strauss, E.G, Strauss, J.H, Baker, T.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2005-03-31 | | Release date: | 2006-02-07 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses.
Structure, 14, 2006
|
|
5ZBR
 
 | | Crystal Structure of Kinesin-3 KIF13B motor domain in AMPPNP form | | Descriptor: | Kinesin family member 13B, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J.Q, Wang, S, Feng, W. | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Delineation of the Neck Linker of Kinesin-3 for Processive Movement.
J. Mol. Biol., 430, 2018
|
|
2MTB
 
 | | Solution structure of apo_FldB | | Descriptor: | Flavodoxin-2 | | Authors: | Jin, C, Fu, W, Ye, Q. | | Deposit date: | 2014-08-15 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Long-chain flavodoxin FldB from Escherichia coli
J.Biomol.Nmr, 60, 2014
|
|
2GQ5
 
 | | Crystal structure of an RNA racemate | | Descriptor: | CALCIUM ION, GLYCEROL, RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), ... | | Authors: | Rypniewski, W, Vallazza, M, Perbandt, M, Klussmann, S, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-04-20 | | Release date: | 2006-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The first crystal structure of an RNA racemate.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
5TO2
 
 | |
3H8Q
 
 | | Crystal structure of glutaredoxin domain of human thioredoxin reductase 3 | | Descriptor: | CHLORIDE ION, SULFATE ION, Thioredoxin reductase 3 | | Authors: | Chaikuad, A, Johansson, C, Ugochukwu, E, Roos, A.K, von Delft, F, Pilka, E, Yue, W, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-29 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of glutaredoxin domain of human thioredoxin reductase 3
To be Published
|
|
2H52
 
 | |
1ZKD
 
 | | X-Ray structure of the putative protein Q6N1P6 from Rhodopseudomonas palustris at the resolution 2.1 A , Northeast Structural Genomics Consortium target RpR58 | | Descriptor: | DUF185 | | Authors: | Kuzin, A.P, Yong, W, Vorobiev, S.M, Acton, T, Ma, L, Xiao, R, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-05-02 | | Release date: | 2005-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-Ray structure of the putative protein Q6N1P6 from Rhodopseudomonas palustris at the resolution 2.1 A , Northeast Structural Genomics Consortium target RpR58
To be Published
|
|
3HBH
 
 | |
2GUZ
 
 | | Structure of the Tim14-Tim16 complex of the mitochondrial protein import motor | | Descriptor: | CITRATE ANION, Mitochondrial import inner membrane translocase subunit TIM14, Mitochondrial import inner membrane translocase subunit TIM16 | | Authors: | Mokranjac, D, Bourenkov, G, Hell, K, Neupert, W, Groll, M. | | Deposit date: | 2006-05-02 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of Tim14 and Tim16, the J and J-like components of the mitochondrial protein import motor.
Embo J., 25, 2006
|
|
2N7Y
 
 | | NMR structure of metal-binding domain 1 of ATP7B | | Descriptor: | Copper-transporting ATPase 2 | | Authors: | Yu, C, Lee, W, Dmitriev, O. | | Deposit date: | 2015-09-27 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of Metal Binding Domain 1 of the Copper Transporter ATP7B Reveals Mechanism of a Singular Wilson Disease Mutation.
Sci Rep, 8, 2018
|
|
3GLP
 
 | | 1.23 A resolution X-ray structure of (GCUGCUGC)2 | | Descriptor: | 5'-R(*GP*CP*UP*GP*CP*UP*GP*C)-3', GLYCEROL, SULFATE ION | | Authors: | Kiliszek, A, Kierzek, R, Krzyzosiak, W.J, Rypniewski, W. | | Deposit date: | 2009-03-12 | | Release date: | 2009-05-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structural insights into CUG repeats containing the 'stretched U-U wobble': implications for myotonic dystrophy.
Nucleic Acids Res., 37, 2009
|
|
5TGP
 
 | | DNA 8mer containing two 2SeT modifications | | Descriptor: | DNA/RNA (5'-R(*G)-D(P*(2ST))-R(P*G)-D(P*(2ST))-R(P*AP*CP*AP*C)-3') | | Authors: | Zhang, W, Huang, Z. | | Deposit date: | 2016-09-28 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA 8mer containing two 2SeT modifications
To Be Published
|
|
1YW6
 
 | | Crystal Structure of Succinylglutamate Desuccinylase from Escherichia coli, Northeast Structural Genomics Target ET72. | | Descriptor: | SULFATE ION, Succinylglutamate desuccinylase | | Authors: | Forouhar, F, Yong, W, Kuzin, A.P, Ciano, M, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-17 | | Release date: | 2005-03-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of Succinylglutamate Desuccinylase from Escherichia coli, Northeast Structural Genomics Target ET72.
To be Published
|
|
2PP0
 
 | | Crystal structure of L-talarate/galactarate dehydratase from Salmonella typhimurium LT2 | | Descriptor: | GLYCEROL, L-talarate/Galactarate Dehydratase | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2007-04-27 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: L-talarate/galactarate dehydratase from Salmonella typhimurium LT2.
Biochemistry, 46, 2007
|
|
2H16
 
 | | Structure of human ADP-ribosylation factor-like 5 (ARL5) | | Descriptor: | ADP-ribosylation factor-like protein 5A, GUANOSINE-5'-DIPHOSPHATE, UNKNOWN ATOM OR ION | | Authors: | Rabeh, W.M, Tempel, W, Yaniw, D, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-16 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human ADP-ribosylation factor-like 5 (ARL5)
To be Published
|
|
2NBI
 
 | | Structure of the PSCD-region of the cell wall protein pleuralin-1 | | Descriptor: | HEP200 protein | | Authors: | De Sanctis, S, Wenzler, M, Kroeger, N, Malloni, W.M, Sumper, M, Rainer, D, Zadravec, P, Brunner, E, Kremer, W, Kalbitzer, H.R. | | Deposit date: | 2016-02-23 | | Release date: | 2016-12-21 | | Method: | SOLUTION NMR | | Cite: | PSCD Domains of Pleuralin-1 from the Diatom Cylindrotheca fusiformis: NMR Structures and Interactions with Other Biosilica-Associated Proteins.
Structure, 24, 2016
|
|
3HJJ
 
 | | Crystal Structure of Maltose O-acetyltransferase from Bacillus anthracis | | Descriptor: | ACETIC ACID, GLYCEROL, Maltose O-acetyltransferase, ... | | Authors: | Kim, Y, Maltseva, N, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-21 | | Release date: | 2009-06-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Crystal Structure of Maltose O-acetyltransferase from Bacillus anthracis
To be Published
|
|
2H3C
 
 | | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA | | Descriptor: | 5'-D(P*AP*TP*AP*TP*GP*TP*AP*TP*AP*CP*CP*CP*G)-3', 5'-D(P*TP*CP*GP*GP*GP*TP*AP*TP*AP*CP*AP*TP*A)-3', CcdA | | Authors: | Madl, T, Van Melderen, L, Respondek, M, Oberer, M, Keller, W, Zangger, K. | | Deposit date: | 2006-05-22 | | Release date: | 2006-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Nucleic Acid and Toxin Recognition of the Bacterial Antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
2H4K
 
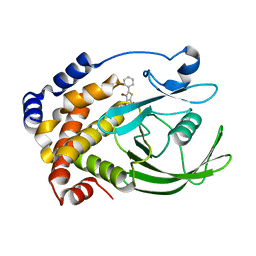 | | Crystal structure of PTP1B with a monocyclic thiophene inhibitor | | Descriptor: | 4-BROMO-3-(CARBOXYMETHOXY)-5-PHENYLTHIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W, Wan, Z.-K. | | Deposit date: | 2006-05-24 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Monocyclic thiophenes as protein tyrosine phosphatase 1B inhibitors: Capturing interactions with Asp48.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2GW2
 
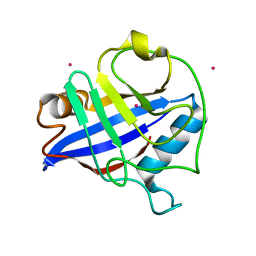 | | Crystal structure of the peptidyl-prolyl isomerase domain of human cyclophilin G | | Descriptor: | Peptidyl-prolyl cis-trans isomerase G, UNKNOWN ATOM OR ION | | Authors: | Bernstein, G, Tempel, W, Davis, T, Newman, E.M, Finerty Jr, P.J, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-03 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and biochemical characterization of the human cyclophilin family of peptidyl-prolyl isomerases.
PLoS Biol., 8, 2010
|
|
2GYP
 
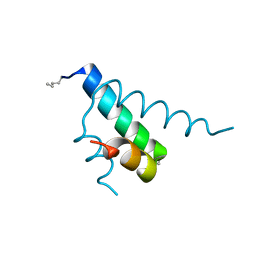 | | Diabetes mellitus due to a frustrated Schellman motif in HNF-1a | | Descriptor: | Hepatocyte nuclear factor 1-alpha | | Authors: | Narayana, N, Phillips, N.B, Hua, Q.X, Jia, W, Weiss, M.A. | | Deposit date: | 2006-05-09 | | Release date: | 2007-03-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diabetes mellitus due to misfolding of a beta-cell transcription factor: stereospecific frustration of a Schellman motif in HNF-1alpha.
J.Mol.Biol., 362, 2006
|
|
1Z8D
 
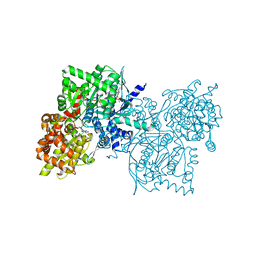 | | Crystal Structure of Human Muscle Glycogen Phosphorylase a with AMP and Glucose | | Descriptor: | ADENINE, ADENOSINE MONOPHOSPHATE, Glycogen phosphorylase, ... | | Authors: | Lukacs, C.M, Oikonomakos, N.G, Crowther, R.L, Hong, L.N, Kammlott, R.U, Levin, W, Li, S, Liu, C.M, Lucas-McGady, D, Pietranico, S, Reik, L. | | Deposit date: | 2005-03-30 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of human muscle glycogen phosphorylase a with bound glucose and AMP: An intermediate conformation with T-state and R-state features.
Proteins, 63, 2006
|
|
