3NIM
 
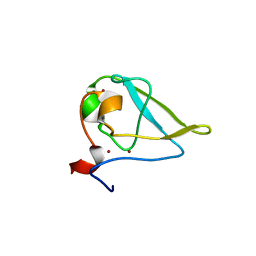 | | The structure of UBR box (RRAA) | | 分子名称: | E3 ubiquitin-protein ligase UBR1, Peptide RRAA, ZINC ION | | 著者 | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | 登録日 | 2010-06-16 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
7FAS
 
 | | VAR2CSA 3D7 ectodomain core region | | 分子名称: | Erythrocyte membrane protein 1, PfEMP1 | | 著者 | Wang, L, Zhaoning, W. | | 登録日 | 2021-07-07 | | 公開日 | 2021-11-17 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | The molecular mechanism of cytoadherence to placental or tumor cells through VAR2CSA from Plasmodium falciparum.
Cell Discov, 7, 2021
|
|
7D9M
 
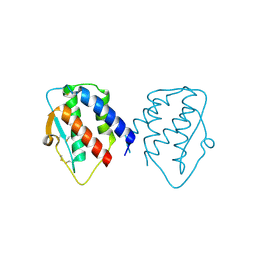 | | grass carp interleukin-2 | | 分子名称: | Interleukin | | 著者 | Junya, w, Jun, z. | | 登録日 | 2020-10-13 | | 公開日 | 2020-10-28 | | 最終更新日 | 2020-11-11 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Structural insights into the co-evolution of IL-2 and its private receptor in fish.
Dev.Comp.Immunol., 115, 2020
|
|
3O3Y
 
 | |
4KGT
 
 | |
1RJ6
 
 | | Crystal Structure of the Extracellular Domain of Murine Carbonic Anhydrase XIV in Complex with Acetazolamide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase XIV, ... | | 著者 | Whittington, D.A, Grubb, J.H, Waheed, A, Shah, G.N, Sly, W.S, Christianson, D.W. | | 登録日 | 2003-11-18 | | 公開日 | 2004-03-09 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Expression, assay, and structure of the extracellular domain of murine carbonic anhydrase XIV: implications for selective inhibition of membrane-associated isozymes.
J.Biol.Chem., 279, 2004
|
|
3O43
 
 | |
7DB6
 
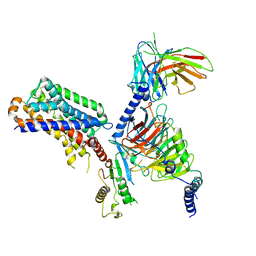 | | human melatonin receptor MT1 - Gi1 complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Okamoto, H.H, Kusakizako, T, Shihioya, W, Yamashita, K, Nishizawa, T, Nureki, O. | | 登録日 | 2020-10-19 | | 公開日 | 2021-08-18 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structure of the human MT 1 -G i signaling complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1U8B
 
 | | Crystal structure of the methylated N-ADA/DNA complex | | 分子名称: | 5'-D(*AP*AP*TP*CP*TP*TP*GP*CP*GP*CP*TP*TP*T)-3', 5'-D(*TP*AP*AP*AP*TP*T)-3', 5'-D(P*AP*AP*AP*GP*CP*GP*CP*AP*AP*GP*AP*T)-3', ... | | 著者 | He, C, Hus, J.-C, Sun, L.J, Zhou, P, Norman, D.P.G, Dotsch, V, Gross, J.D, Lane, W.S, Wagner, G, Verdine, G.L. | | 登録日 | 2004-08-05 | | 公開日 | 2005-10-11 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A methylation-dependent electrostatic switch controls DNA repair and transcriptional activation by E. coli ada.
Mol.Cell, 20, 2005
|
|
4NJ0
 
 | | GCN4-p1 single Val9 to Ile mutant | | 分子名称: | General control protein GCN4 | | 著者 | Oshaben, K.M, Horne, W.S. | | 登録日 | 2013-11-08 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Tuning assembly size in Peptide-based supramolecular polymers by modulation of subunit association affinity.
Biomacromolecules, 15, 2014
|
|
4NJ1
 
 | | GCN4-p1 double Val9, 23 to Ile mutant | | 分子名称: | General control protein GCN4 | | 著者 | Oshaben, K.M, Horne, W.S. | | 登録日 | 2013-11-08 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Tuning assembly size in Peptide-based supramolecular polymers by modulation of subunit association affinity.
Biomacromolecules, 15, 2014
|
|
3NIH
 
 | | The structure of UBR box (RIAAA) | | 分子名称: | E3 ubiquitin-protein ligase UBR1, Peptide RIAAA, ZINC ION | | 著者 | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | 登録日 | 2010-06-16 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
1SO4
 
 | | Crystal structure of K64A mutant of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-threonohydroxamate 4-phosphate | | 分子名称: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-THREONOHYDROXAMATE 4-PHOSPHATE, MAGNESIUM ION | | 著者 | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | 登録日 | 2004-03-12 | | 公開日 | 2004-06-08 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Evolution of Enzymatic Activities in the Orotidine 5'-Monophosphate Decarboxylase Suprafamily: Crystallographic Evidence for a Proton Relay System in the Active Site of 3-Keto-l-gulonate 6-Phosphate Decarboxylase(,)
Biochemistry, 43, 2004
|
|
1SHC
 
 | | SHC PTB DOMAIN COMPLEXED WITH A TRKA RECEPTOR PHOSPHOPEPTIDE, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | SHC, TRKA RECEPTOR PHOSPHOPEPTIDE | | 著者 | Zhou, M.-M, Ravichandran, K.S, Olejniczak, E.T, Petros, A.M, Meadows, R.P, Sattler, M, Harlan, J.E, Wade, W.S, Burakoff, S.J, Fesik, S.W. | | 登録日 | 1996-03-27 | | 公開日 | 1997-05-15 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and ligand recognition of the phosphotyrosine binding domain of Shc.
Nature, 378, 1995
|
|
1SQ0
 
 | | Crystal Structure of the Complex of the Wild-type Von Willebrand Factor A1 domain and Glycoprotein Ib alpha at 2.6 Angstrom Resolution | | 分子名称: | Platelet glycoprotein Ib alpha chain (Glycoprotein Ibalpha) (GP-Ib alpha) (GPIbA) (GPIb-alpha) (CD42B-alpha) (CD42B) [Contains: Glycocalicin], Von Willebrand factor (vWF) [Contains: Von Willebrand antigen II] | | 著者 | Dumas, J.J, Kumar, R, McDonagh, T, Sullivan, F, Stahl, M.L, Somers, W.S, Mosyak, L. | | 登録日 | 2004-03-17 | | 公開日 | 2004-04-13 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of the wild-type von Willebrand factor A1-glycoprotein Ibalpha complex reveals conformation differences with a complex bearing von Willebrand disease mutations
J.Biol.Chem., 279, 2004
|
|
4NIZ
 
 | |
4NJ2
 
 | | GCN4-p1 triple Val9, 23,30 to Ile mutant | | 分子名称: | GLYCEROL, General control protein GCN4 | | 著者 | Oshaben, K.M, Horne, W.S. | | 登録日 | 2013-11-08 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Tuning assembly size in Peptide-based supramolecular polymers by modulation of subunit association affinity.
Biomacromolecules, 15, 2014
|
|
3OY3
 
 | | Crystal structure of ABL T315I mutant kinase domain bound with a DFG-out inhibitor AP24589 | | 分子名称: | 5-[(5-{[4-{[4-(2-hydroxyethyl)piperazin-1-yl]methyl}-3-(trifluoromethyl)phenyl]carbamoyl}-2-methylphenyl)ethynyl]-1-methyl-1H-imidazole-2-carboxamide, Tyrosine-protein kinase ABL1 | | 著者 | Zhou, T, Commodore, L, Huang, W.S, Wang, Y, Thomas, M, Keats, J, Xu, Q, Rivera, V, Shakespeare, W.C, Clackson, T, Dalgarno, D.C, Zhu, X. | | 登録日 | 2010-09-22 | | 公開日 | 2010-12-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural Mechanism of the Pan-BCR-ABL Inhibitor Ponatinib (AP24534): Lessons for Overcoming Kinase Inhibitor Resistance.
Chem.Biol.Drug Des., 77, 2011
|
|
1SO3
 
 | | Crystal structure of H136A mutant of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-threonohydroxamate 4-phosphate | | 分子名称: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-THREONOHYDROXAMATE 4-PHOSPHATE, MAGNESIUM ION | | 著者 | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | 登録日 | 2004-03-12 | | 公開日 | 2004-06-08 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Evolution of Enzymatic Activities in the Orotidine 5'-Monophosphate Decarboxylase Suprafamily: Crystallographic Evidence for a Proton Relay System in the Active Site of 3-Keto-l-gulonate 6-Phosphate Decarboxylase(,)
Biochemistry, 43, 2004
|
|
1SO5
 
 | | Crystal structure of E112Q mutant of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-threonohydroxamate 4-phosphate | | 分子名称: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-THREONOHYDROXAMATE 4-PHOSPHATE, MAGNESIUM ION | | 著者 | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | 登録日 | 2004-03-12 | | 公開日 | 2004-06-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Evolution of Enzymatic Activities in the Orotidine 5'-Monophosphate Decarboxylase Suprafamily: Crystallographic Evidence for a Proton Relay System in the Active Site of 3-Keto-l-gulonate 6-Phosphate Decarboxylase(,)
Biochemistry, 43, 2004
|
|
7FJB
 
 | | KpAckA (PduW) with AMPPNP, sodium acetate complex structure | | 分子名称: | ACETATE ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Probable propionate kinase, ... | | 著者 | Wenyue, W, Zhang, Q, Bartlam, M. | | 登録日 | 2021-08-03 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | KpAckA (PduW) with AMPPNP, sodium acetate complex structure
To Be Published
|
|
4KCB
 
 | | Crystal Structure of Exo-1,5-alpha-L-arabinanase from Bovine Ruminal Metagenomic Library | | 分子名称: | Arabinan endo-1,5-alpha-L-arabinosidase, PHOSPHATE ION | | 著者 | Santos, C.R, Polo, C.C, Costa, M.C.M.F, Nascimento, A.F.Z, Wong, D.W.S, Murakami, M.T. | | 登録日 | 2013-04-24 | | 公開日 | 2014-02-05 | | 最終更新日 | 2014-04-09 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Mechanistic strategies for catalysis adopted by evolutionary distinct family 43 arabinanases.
J.Biol.Chem., 289, 2014
|
|
3OXZ
 
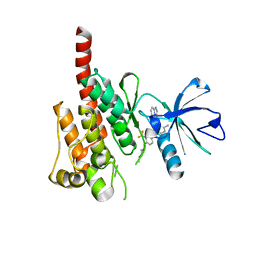 | | Crystal structure of ABL kinase domain bound with a DFG-out inhibitor AP24534 | | 分子名称: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Tyrosine-protein kinase ABL1 | | 著者 | Zhou, T, Huang, W.S, Wang, Y, Thomas, M, Keats, J, Xu, Q, Rivera, V, Shakespeare, W.C, Clackson, T, Dalgarno, D.C, Zhu, X. | | 登録日 | 2010-09-22 | | 公開日 | 2010-12-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Mechanism of the Pan-BCR-ABL Inhibitor Ponatinib (AP24534): Lessons for Overcoming Kinase Inhibitor Resistance.
Chem.Biol.Drug Des., 77, 2011
|
|
4NX9
 
 | |
4NJ4
 
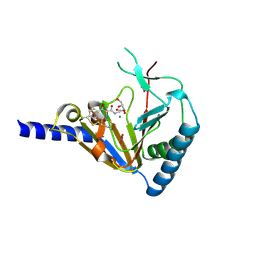 | | Crystal Structure of Human ALKBH5 | | 分子名称: | GLYCEROL, MANGANESE (II) ION, N-[(1-CHLORO-4-HYDROXYISOQUINOLIN-3-YL)CARBONYL]GLYCINE, ... | | 著者 | Aik, W.S, McDonough, M.A, Schofield, C.J. | | 登録日 | 2013-11-08 | | 公開日 | 2014-01-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Structure of human RNA N6-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation.
Nucleic Acids Res., 42, 2014
|
|
