7GH2
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-090737b9-1 (Mpro-x12735) | | 分子名称: | (4R)-6-chloro-N-(isoquinolin-4-yl)-N-propanoyl-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4E74
 
 | | Crystal structure of the RHO GTPASE BINDING DOMAIN of Plexin A4A | | 分子名称: | Plexin-A4, UNKNOWN ATOM OR ION | | 著者 | Guan, X, Wang, H, Tempel, W, Dong, A, Tong, Y, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | 登録日 | 2012-03-16 | | 公開日 | 2012-03-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Crystal structure of the RHO GTPASE BINDING DOMAIN of Plexin A4A
to be published
|
|
7GH4
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-00c1612e-1 (Mpro-x12777) | | 分子名称: | 2-(3-chlorophenyl)-N-(6-methoxyisoquinolin-4-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GH5
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with TRY-UNI-714-12 (Mpro-x2908) | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(3-chlorophenyl)-N'-(4-methylpyridin-3-yl)urea | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4W87
 
 | | Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from metagenomic library, in complex with a xyloglucan oligosaccharide | | 分子名称: | MAGNESIUM ION, Xyloglucan-specific endo-beta-1,4-glucanase, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | 登録日 | 2014-08-22 | | 公開日 | 2015-03-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
6L13
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | 分子名称: | 2-chloranyl-10-(2-piperidin-4-ylethyl)phenoxazine, Serine/threonine-protein kinase pim-1 | | 著者 | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | 登録日 | 2019-09-27 | | 公開日 | 2020-05-27 | | 最終更新日 | 2020-07-08 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L2L
 
 | |
5WYG
 
 | |
4ED2
 
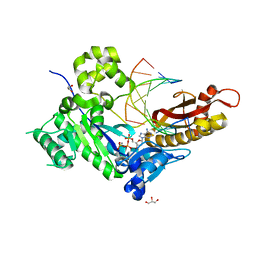 | | Human DNA polymerase eta - DNA ternary complex: AT crystal at pH 7.2 (Na+ HEPES) with 1 Ca2+ ion | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | 著者 | Nakamura, T, Zhao, Y, Yang, W. | | 登録日 | 2012-03-26 | | 公開日 | 2012-07-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.711 Å) | | 主引用文献 | Watching DNA polymerase eta make a phosphodiester bond
Nature, 487, 2012
|
|
5WZ2
 
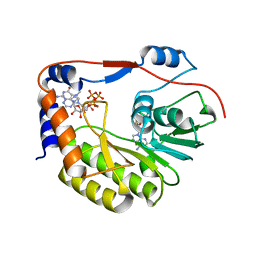 | | Crystal structure of Zika virus NS5 methyltransferase bound to SAM and RNA analogue (m7GpppA) | | 分子名称: | NS5 MTase, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, S-ADENOSYLMETHIONINE | | 著者 | Duan, W, Song, H, Qi, J, Shi, Y, Gao, G.F. | | 登録日 | 2017-01-16 | | 公開日 | 2017-03-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The crystal structure of Zika virus NS5 reveals conserved drug targets.
EMBO J., 36, 2017
|
|
4ECU
 
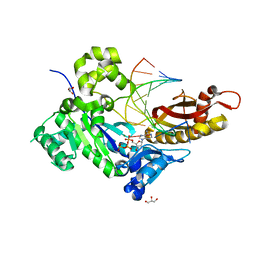 | | Human DNA polymerase eta - DNA ternary complex: Reaction in the AT crystal at pH 7.0 for 200 sec | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | 著者 | Nakamura, T, Zhao, Y, Yang, W. | | 登録日 | 2012-03-26 | | 公開日 | 2012-07-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.949 Å) | | 主引用文献 | Watching DNA polymerase eta make a phosphodiester bond
Nature, 487, 2012
|
|
4ED6
 
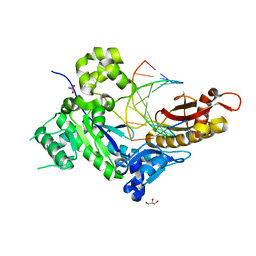 | | Human DNA polymerase eta - DNA ternary complex: Reaction in the AT crystal at pH 6.7 for 15 hr, Sideway translocation | | 分子名称: | DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*TP*A)-3'), DNA (5'-D(*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta, ... | | 著者 | Nakamura, T, Zhao, Y, Yang, W. | | 登録日 | 2012-03-27 | | 公開日 | 2012-07-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.211 Å) | | 主引用文献 | Watching DNA polymerase eta make a phosphodiester bond
Nature, 487, 2012
|
|
1DKF
 
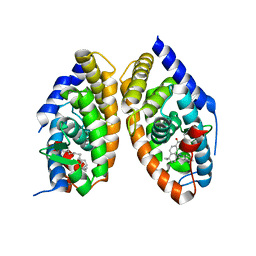 | | CRYSTAL STRUCTURE OF A HETERODIMERIC COMPLEX OF RAR AND RXR LIGAND-BINDING DOMAINS | | 分子名称: | 4-[(4,4-DIMETHYL-1,2,3,4-TETRAHYDRO-[1,2']BINAPTHALENYL-7-CARBONYL)-AMINO]-BENZOIC ACID, OLEIC ACID, PROTEIN (RETINOIC ACID RECEPTOR-ALPHA), ... | | 著者 | Bourguet, W, Vivat, V, Wurtz, J.M, Chambon, P, Gronemeyer, H, Moras, D, Structural Proteomics in Europe (SPINE) | | 登録日 | 1999-12-07 | | 公開日 | 2000-04-19 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains.
Mol.Cell, 5, 2000
|
|
6ACK
 
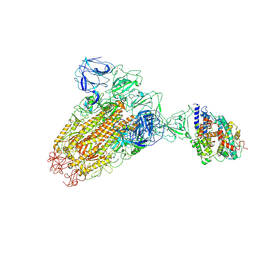 | |
1SMO
 
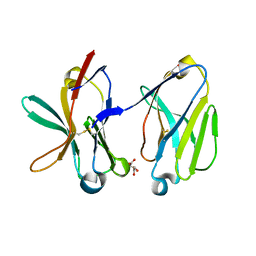 | | Crystal Structure of Human Triggering Receptor Expressed on Myeloid Cells 1 (TREM-1) at 1.47 . | | 分子名称: | L(+)-TARTARIC ACID, triggering receptor expressed on myeloid cells 1 | | 著者 | Kelker, M.S, Foss, T.R, Peti, W, Teyton, L, Kelly, J.W, Wilson, I.A. | | 登録日 | 2004-03-09 | | 公開日 | 2004-09-21 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Crystal Structure of Human Triggering Receptor Expressed on Myeloid Cells 1 (TREM-1) at 1.47A.
J.Mol.Biol., 342, 2004
|
|
1DLB
 
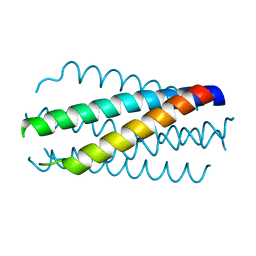 | | HELICAL INTERACTIONS IN THE HIV-1 GP41 CORE REVEALS STRUCTURAL BASIS FOR THE INHIBITORY ACTIVITY OF GP41 PEPTIDES | | 分子名称: | HIV-1 ENVELOPE GLYCOPROTEIN GP41 | | 著者 | Shu, W, Liu, J, Ji, H, Rading, L, Jiang, S, Lu, M. | | 登録日 | 1999-12-09 | | 公開日 | 1999-12-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Helical interactions in the HIV-1 gp41 core reveal structural basis for the inhibitory activity of gp41 peptides.
Biochemistry, 39, 2000
|
|
4EGG
 
 | | Computationally Designed Self-assembling tetrahedron protein, T310 | | 分子名称: | GLYCEROL, Putative acetyltransferase SACOL2570 | | 著者 | Sawaya, M.R, King, N.P, Sheffler, W, Baker, D, Yeates, T.O. | | 登録日 | 2012-03-30 | | 公開日 | 2012-05-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Computational design of self-assembling protein nanomaterials with atomic level accuracy.
Science, 336, 2012
|
|
1DG0
 
 | | NMR STRUCTURE OF DES[GLY1]-CONTRYPHAN-R CYCLIC PEPTIDE (MAJOR FORM) | | 分子名称: | DES[GLY1]-CONTRYPHAN-R | | 著者 | Pallaghy, P.K, He, W, Jimenez, E.C, Olivera, B.M, Norton, R.S. | | 登録日 | 1999-11-22 | | 公開日 | 2003-09-09 | | 最終更新日 | 2020-06-24 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structures of the contryphan family of cyclic peptides. Role of electrostatic interactions in cis-trans isomerism
Biochemistry, 39, 2000
|
|
5WZ3
 
 | | Crystal structure of Zika virus NS5 RNA-dependent RNA polymerase(RdRP) | | 分子名称: | NS5 RdRp, ZINC ION | | 著者 | Duan, W, Song, H, Qi, J, Shi, Y, Gao, G.F. | | 登録日 | 2017-01-16 | | 公開日 | 2017-03-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.804 Å) | | 主引用文献 | The crystal structure of Zika virus NS5 reveals conserved drug targets.
EMBO J., 36, 2017
|
|
5X29
 
 | |
1DCH
 
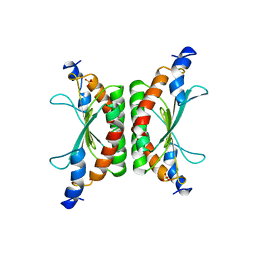 | | CRYSTAL STRUCTURE OF DCOH, A BIFUNCTIONAL, PROTEIN-BINDING TRANSCRIPTION COACTIVATOR | | 分子名称: | DCOH (DIMERIZATION COFACTOR OF HNF-1), SULFATE ION | | 著者 | Endrizzi, J.A, Cronk, J.D, Wang, W, Crabtree, G.R, Alber, T. | | 登録日 | 1995-01-24 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of DCoH, a bifunctional, protein-binding transcriptional coactivator.
Science, 268, 1995
|
|
1SOL
 
 | | A PIP2 AND F-ACTIN-BINDING SITE OF GELSOLIN, RESIDUE 150-169 (NMR, AVERAGED STRUCTURE) | | 分子名称: | GELSOLIN (150-169) | | 著者 | Xian, W, Vegners, R, Janmey, P.A, Braunlin, W.H. | | 登録日 | 1995-09-29 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Spectroscopic studies of a phosphoinositide-binding peptide from gelsolin: behavior in solutions of mixed solvent and anionic micelles.
Biophys.J., 69, 1995
|
|
1DFF
 
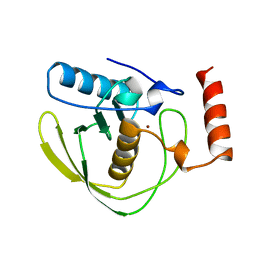 | | PEPTIDE DEFORMYLASE | | 分子名称: | PEPTIDE DEFORMYLASE, ZINC ION | | 著者 | Chan, M.K, Gong, W, Rajagopalan, P.T.R, Hao, B, Tsai, C.M, Pei, D. | | 登録日 | 1997-08-19 | | 公開日 | 1998-09-02 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | Crystal structure of the Escherichia coli peptide deformylase.
Biochemistry, 36, 1997
|
|
4EP1
 
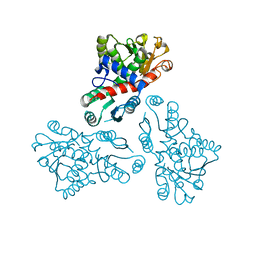 | | Crystal structure of anabolic ornithine carbamoyltransferase from Bacillus anthracis | | 分子名称: | Ornithine carbamoyltransferase | | 著者 | Shabalin, I.G, Mikolajczak, K, Stam, J, Winsor, J, Shuvalova, L, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-04-16 | | 公開日 | 2012-04-25 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Crystal structures of anabolic ornithine carbamoyltransferase from Bacillus anthracis
To be Published
|
|
6L14
 
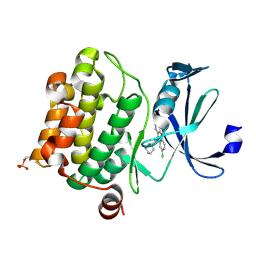 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | 分子名称: | 2-chloranyl-10-[3-[(3~{S})-piperidin-3-yl]propyl]phenoxazine, Serine/threonine-protein kinase pim-1 | | 著者 | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | 登録日 | 2019-09-27 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
